| Entry |
|
|---|
| Name |
Alanine, aspartate and glutamate metabolism - Mycobacterium canettii CIPT 140070008 |
|---|
| Class |
Metabolism; Amino acid metabolism
|
|---|
| Pathway map |
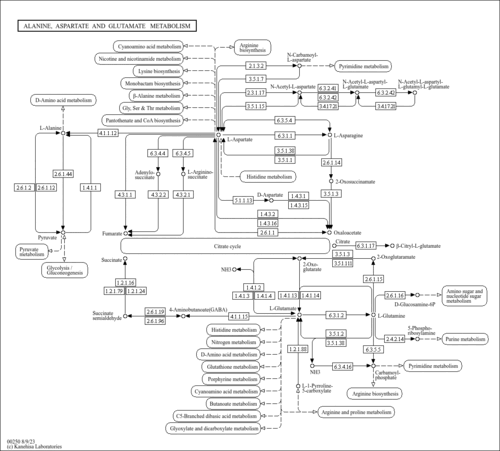
| mcv00250 | Alanine, aspartate and glutamate metabolism |
|
|---|
| Module |
|
|---|
| Other DBs |
|
|---|
| Organism |
Mycobacterium canettii CIPT 140070008 [GN: mcv]
|
|---|
| Gene |
| BN43_10388 | purA; Putative adenylosuccinate synthetase PurA (IMP--aspartate ligase) (ADSS) (AMPsase) [KO:K01939] [EC:6.3.4.4] |
| BN43_30485 | pyrB; Putative aspartate carbamoyltransferase PyrB (ATCase) (aspartate transcarbamylase) [KO:K00609] [EC:2.1.3.2] |
| BN43_40264 | gabT; 4-aminobutyrate aminotransferase GabT (gamma-amino-N-butyrate transaminase) (GABA transaminase) (glutamate:succinic semialdehyde transaminase) (GABA aminotransferase) (GABA-AT) [KO:K07250] [EC:2.6.1.19 2.6.1.22 2.6.1.48] |
| BN43_90390 | gltB; Putative ferredoxin-dependent glutamate synthase [NADPH] (large subunit) GltB (l-glutamate synthase) (l-glutamate synthetase) (NADH-glutamate synthase) (glutamate synthase (NADH))(NADPH-GOGAT) [KO:K00265] [EC:1.4.1.13] |
| BN43_90389 | gltD; Putative NADH-dependent glutamate synthase (small subunit) GltD (l-glutamate synthase) (l-glutamate synthetase) (NADH-glutamate synthase) (glutamate synthase (NADH)) (GltS beta chain) (NADPH-GOGAT) [KO:K00266] [EC:1.4.1.13] |
| BN43_40139 | gdh; Putative NAD-dependent glutamate dehydrogenase Gdh (NAD-GDH) (NAD-dependent glutamic dehydrogenase) [KO:K15371] [EC:1.4.1.2] |
| BN43_30491 | carB; Carbamoyl-phosphate synthase large chain CarB (carbamoyl-phosphate synthetase ammonia chain) [KO:K01955] [EC:6.3.5.5] |
| BN43_30488 | carA; Putative carbamoyl-phosphate synthase small chain CarA (carbamoyl-phosphate synthetase glutamine chain) [KO:K01956] [EC:6.3.5.5] |
| BN43_70104 | glmS; Putative Glucosamine--Fructose-6-Phosphate aminotransferase GlmS (Hexosephosphate Aminotransferase) (D-Fructose-6-Phosphate Aminotransferase) (GfaT) (L-Glutamine-D-Fructose-6-Phosphate Aminotransferase) (Glucosamine-6-Phosphate Synthase) [KO:K00820] [EC:2.6.1.16] |
| BN43_20250 | purF; Amidophosphoribosyltransferase PurF (glutamine phosphoribosylpyrophosphate amidotransferase) (ATASE) (GPATASE) [KO:K00764] [EC:2.4.2.14] |
|
|---|
| Compound |
| C00352 | D-Glucosamine 6-phosphate |
| C00438 | N-Carbamoyl-L-aspartate |
| C03406 | N-(L-Arginino)succinate |
| C03794 | N6-(1,2-Dicarboxyethyl)-AMP |
| C03912 | (S)-1-Pyrroline-5-carboxylate |
| C12270 | N-Acetylaspartylglutamate |
| C20775 | beta-Citryl-L-glutamate |
| C20776 | N-Acetylaspartylglutamylglutamate |
|
|---|
| Reference |
|
|---|
| Authors |
Nishizuka Y, Seyama Y, Ikai A, Ishimura Y, Kawaguchi A (eds). |
|---|
| Title |
[Cellular Functions and Metabolic Maps] (In Japanese) |
|---|
| Journal |
Tokyo Kagaku Dojin (1997) |
|---|
| Reference |
|
|---|
| Authors |
Wu G |
|---|
| Title |
Intestinal mucosal amino acid catabolism. |
|---|
| Journal |
|
|---|
Related
pathway |
| mcv00260 | Glycine, serine and threonine metabolism |
| mcv00330 | Arginine and proline metabolism |
| mcv00520 | Amino sugar and nucleotide sugar metabolism |
| mcv00630 | Glyoxylate and dicarboxylate metabolism |
| mcv00660 | C5-Branched dibasic acid metabolism |
| mcv00760 | Nicotinate and nicotinamide metabolism |
| mcv00770 | Pantothenate and CoA biosynthesis |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|