| Entry |
|
|---|
| Name |
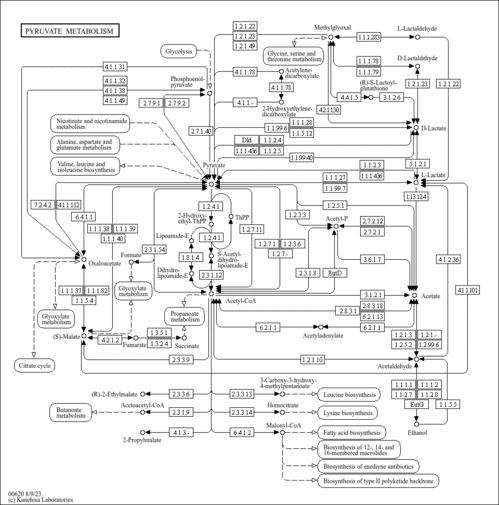
Pyruvate metabolism - Mycobacterium tuberculosis 7199-99 |
|---|
| Class |
Metabolism; Carbohydrate metabolism
|
|---|
| Pathway map |
|
|---|
| Module |
| mtub_M00579 | Phosphate acetyltransferase-acetate kinase pathway, acetyl-CoA => acetate [PATH:mtub00620] |
|
|---|
| Other DBs |
|
|---|
| Organism |
Mycobacterium tuberculosis 7199-99 [GN: mtub]
|
|---|
| Gene |
| MT7199_3730 | ACETYL-COENZYME A SYNTHETASE ACS (ACETATE-CoA LIGASE) (ACETYL-CoA SYNTHETASE) (ACETYL-CoA SYNTHASE) (ACYL-ACTIVATING ENZYME) (ACETATE THIOKINASE) (ACETYL-ACTIVATING ENZYME) (ACETATE-COENZYME A LIGASE) (ACETYL-COENZYME A SYNTHASE) [KO:K01895] [EC:6.2.1.1] |
| MT7199_2273 | putative pyruvate dehydrogenase E1 component aceE (PYRUVATE DECARBOXYLASE) (PYRUVATE DEHYDROGENASE) (PYRUVIC DEHYDROGENASE) [KO:K00163] [EC:1.2.4.1] |
| MT7199_0471 | lpdC; DIHYDROLIPOAMIDE DEHYDROGENASE LPD (LIPOAMIDE REDUCTASE (NADH)) (LIPOYL DEHYDROGENASE) (DIHYDROLIPOYL DEHYDROGENASE) (DIAPHORASE) [KO:K00382] [EC:1.8.1.4] |
| MT7199_3327 | putative BIFUNCTIONAL protein ACETYL-/PROPIONYL-COENZYME A CARBOXYLASE (ALPHA CHAIN) ACCA3: BIOTIN CARBOXYLASE + BIOTIN CARBOXYL CARRIER protein (BCCP) [KO:K11263] [EC:6.4.1.2 6.4.1.3 6.4.1.- 6.3.4.14] |
| MT7199_2363 | putative [NAD] DEPENDENT MALATE OXIDOREDUCTASE MEZ (MALIC ENZYME) (NAD-MALIC ENZYME) (MALATE DEHYDROGENASE (OXALOACETATE DECARBOXYLATING)) (PYRUVIC-MALIC CARBOXYLASE) (NAD-ME) [KO:K00027] [EC:1.1.1.38] |
| MT7199_1588 | putative FUMARATE REDUCTASE [FLAVOprotein SUBUNIT] FRDA (FUMARATE DEHYDROGENASE) (FUMARIC HYDROGENASE) [KO:K00244] [EC:1.3.5.1] |
| MT7199_1589 | putative FUMARATE REDUCTASE [IRON-SULFUR SUBUNIT] FRDB (FUMARATE DEHYDROGENASE) (FUMARIC HYDROGENASE) [KO:K00245] [EC:1.3.5.1] |
| MT7199_1590 | putative FUMARATE REDUCTASE [MEMBRANE ANCHOR SUBUNIT] FRDC (FUMARATE DEHYDROGENASE) (FUMARIC HYDROGENASE) [KO:K00246] |
| MT7199_1591 | putative FUMARATE REDUCTASE [MEMBRANE ANCHOR SUBUNIT] FRDD (FUMARATE DEHYDROGENASE) (FUMARIC HYDROGENASE) [KO:K00247] |
| MT7199_0215 | putative IRON-REGULATED PHOSPHOENOLPYRUVATE CARBOXYKINASE [GTP] PCKA (PHOSPHOENOLPYRUVATE CARBOXYLASE) (PEPCK)(PEP CARBOXYKINASE) [KO:K01596] [EC:4.1.1.32] |
| MT7199_0248 | putative ACETYL-CoA ACYLTRANSFERASE FADA2 (3-KETOACYL-CoA THIOLASE) (BETA-KETOTHIOLASE) [KO:K00626] [EC:2.3.1.9] |
| MT7199_3776 | leuA; 2-ISOPROPYLMALATE SYNTHASE LEUA (ALPHA-ISOPROPYLMALATE SYNTHASE) (ALPHA-IPM SYNTHETASE) [KO:K01649] [EC:2.3.3.13] |
|
|---|
| Compound |
| C01251 | (R)-2-Hydroxybutane-1,2,4-tricarboxylate |
| C03451 | (R)-S-Lactoylglutathione |
| C03981 | 2-Hydroxyethylenedicarboxylate |
| C05125 | 2-(alpha-Hydroxyethyl)thiamine diphosphate |
| C15972 | Enzyme N6-(lipoyl)lysine |
| C15973 | Enzyme N6-(dihydrolipoyl)lysine |
| C16255 | [Dihydrolipoyllysine-residue acetyltransferase] S-acetyldihydrolipoyllysine |
|
|---|
| Reference |
|
|---|
| Authors |
Nishizuka Y, Seyama Y, Ikai A, Ishimura Y, Kawaguchi A (eds). |
|---|
| Title |
[Cellular Functions and Metabolic Maps] (In Japanese) |
|---|
| Journal |
Tokyo Kagaku Dojin (1997) |
|---|
Related
pathway |
| mtub00250 | Alanine, aspartate and glutamate metabolism |
| mtub00260 | Glycine, serine and threonine metabolism |
| mtub00290 | Valine, leucine and isoleucine biosynthesis |
| mtub00630 | Glyoxylate and dicarboxylate metabolism |
| mtub00760 | Nicotinate and nicotinamide metabolism |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|