 | | PATHWAY: ko00620 | |
| Entry |
|
|---|
| Name |
Pyruvate metabolism |
|---|
| Class |
Metabolism; Carbohydrate metabolism
|
|---|
| Pathway map |
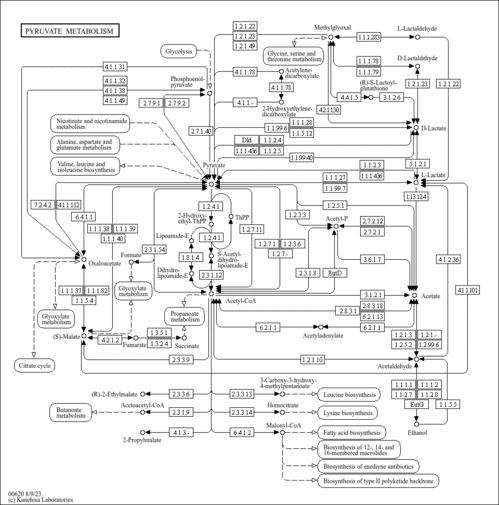
|
|---|
| Module |
| M00172 | C4-dicarboxylic acid cycle, NADP - malic enzyme type [PATH:ko00620] |
| M00579 | Phosphate acetyltransferase-acetate kinase pathway, acetyl-CoA => acetate [PATH:ko00620] |
|
|---|
| Other DBs |
|
|---|
| Orthology |
| K00027 | ME2, sfcA, maeA; malate dehydrogenase (oxaloacetate-decarboxylating) [EC:1.1.1.38] |
| K00028 | E1.1.1.39; malate dehydrogenase (decarboxylating) [EC:1.1.1.39] |
| K00029 | maeB; malate dehydrogenase (oxaloacetate-decarboxylating)(NADP+) [EC:1.1.1.40] |
| K00102 | LDHD, dld; D-lactate dehydrogenase (cytochrome) [EC:1.1.2.4] |
| K00121 | frmA, ADH5, adhC; S-(hydroxymethyl)glutathione dehydrogenase / alcohol dehydrogenase [EC:1.1.1.284 1.1.1.1] |
| K00132 | E1.2.1.10; acetaldehyde dehydrogenase (acetylating) [EC:1.2.1.10] |
| K00138 | aldB; aldehyde dehydrogenase [EC:1.2.1.-] |
| K00161 | PDHA; pyruvate dehydrogenase E1 component subunit alpha [EC:1.2.4.1] |
| K00162 | PDHB; pyruvate dehydrogenase E1 component subunit beta [EC:1.2.4.1] |
| K00169 | porA; pyruvate ferredoxin oxidoreductase alpha subunit [EC:1.2.7.1] |
| K00170 | porB; pyruvate ferredoxin oxidoreductase beta subunit [EC:1.2.7.1] |
| K00171 | porD; pyruvate ferredoxin oxidoreductase delta subunit [EC:1.2.7.1] |
| K00172 | porC, porG; pyruvate ferredoxin oxidoreductase gamma subunit [EC:1.2.7.1] |
| K00174 | korA, oorA, oforA; 2-oxoglutarate/2-oxoacid ferredoxin oxidoreductase subunit alpha [EC:1.2.7.3 1.2.7.11] |
| K00175 | korB, oorB, oforB; 2-oxoglutarate/2-oxoacid ferredoxin oxidoreductase subunit beta [EC:1.2.7.3 1.2.7.11] |
| K00189 | vorG, porG; 2-oxoisovalerate/pyruvate ferredoxin oxidoreductase gamma subunit [EC:1.2.7.7 1.2.7.1] |
| K00233 | frdA, fdrA; succinate dehydrogenase flavoprotein subunit |
| K00244 | frdA; succinate dehydrogenase flavoprotein subunit [EC:1.3.5.1] |
| K00245 | frdB; succinate dehydrogenase iron-sulfur subunit [EC:1.3.5.1] |
| K00246 | frdC; succinate dehydrogenase subunit C |
| K00247 | frdD; succinate dehydrogenase subunit D |
| K00627 | DLAT, aceF, pdhC; pyruvate dehydrogenase E2 component (dihydrolipoyllysine-residue acetyltransferase) [EC:2.3.1.12] |
| K01069 | gloB, gloC, HAGH; hydroxyacylglutathione hydrolase [EC:3.1.2.6] |
| K01571 | oadA; oxaloacetate decarboxylase (Na+ extruding) subunit alpha [EC:7.2.4.2] |
| K01573 | oadG; oxaloacetate decarboxylase (Na+ extruding) subunit gamma |
| K01596 | E4.1.1.32, pckA, PCK; phosphoenolpyruvate carboxykinase (GTP) [EC:4.1.1.32] |
| K01675 | fumD; fumarate hydratase D |
| K01676 | E4.2.1.2A, fumA, fumB; fumarate hydratase, class I [EC:4.2.1.2] |
| K01677 | E4.2.1.2AA, fumA; fumarate hydratase subunit alpha [EC:4.2.1.2] |
| K01678 | E4.2.1.2AB, fumB; fumarate hydratase subunit beta [EC:4.2.1.2] |
| K01679 | E4.2.1.2B, fumC, FH; fumarate hydratase, class II [EC:4.2.1.2] |
| K01774 | fumE; fumarate hydratase E |
| K01905 | acdA; acetate---CoA ligase (ADP-forming) subunit alpha [EC:6.2.1.13] |
| K02160 | accB, bccP; acetyl-CoA carboxylase biotin carboxyl carrier protein |
| K04020 | eutD; phosphotransacetylase |
| K04021 | eutE; aldehyde dehydrogenase |
| K04022 | eutG; alcohol dehydrogenase |
| K11263 | bccA, pccA, accA3; acetyl-CoA/propionyl-CoA/long-chain acyl-CoA carboxylase, biotin carboxylase, biotin carboxyl carrier protein [EC:6.4.1.2 6.4.1.3 6.4.1.- 6.3.4.14] |
| K13953 | adhP; alcohol dehydrogenase, propanol-preferring [EC:1.1.1.1] |
| K14028 | mdh1, mxaF; methanol dehydrogenase (cytochrome c) subunit 1 [EC:1.1.2.7] |
| K14029 | mdh2, mxaI; methanol dehydrogenase (cytochrome c) subunit 2 [EC:1.1.2.7] |
| K18930 | dld; D-lactate dehydrogenase |
| K20370 | PEPCK; phosphoenolpyruvate carboxykinase (diphosphate) [EC:4.1.1.38] |
| K20509 | madB, oadB, gcdB, mmdB; carboxybiotin decarboxylase [EC:7.2.4.1] |
| K21618 | DLD3; (R)-2-hydroxyglutarate---pyruvate transhydrogenase [EC:1.1.99.40] |
| K22224 | acdB; acetate---CoA ligase (ADP-forming) subunit beta [EC:6.2.1.13] |
| K22473 | adhA; alcohol dehydrogenase (quinone), dehydrogenase subunit [EC:1.1.5.5] |
| K22474 | adhB; alcohol dehydrogenase (quinone), cytochrome c subunit [EC:1.1.5.5] |
| K23257 | yvgN; methylglyoxal/glyoxal reductase |
| K24012 | acdAB; acetate---CoA ligase (ADP-forming) |
| K25995 | frdB, fdrB; succinate dehydrogenase iron-sulfur subunit |
| K25996 | frdC, fdrC; succinate dehydrogenase subunit C |
| K26113 | lctD; lactate dehydrogenase (NAD+,ferredoxin) subunit LctD |
| K26114 | lctC; lactate dehydrogenase (NAD+,ferredoxin) subunit LctC |
| K26115 | lctB; lactate dehydrogenase (NAD+,ferredoxin) subunit LctB |
| K26318 | fccA, fcc3; fumarate reductase (cytochrome) |
| K28615 | adh; alcohol / (R,R)-butanediol / aldehyde dehydrogenase |
| K28726 | porA; pyruvate synthase (flavodoxin) subunit A |
| K28727 | porB; pyruvate synthase (flavodoxin) subunit B |
| K28728 | porC; pyruvate synthase (flavodoxin) subunit C |
| K28729 | porD; pyruvate synthase (flavodoxin) subunit D |
|
|---|
| Compound |
| C01251 | (R)-2-Hydroxybutane-1,2,4-tricarboxylate |
| C03451 | (R)-S-Lactoylglutathione |
| C03981 | 2-Hydroxyethylenedicarboxylate |
| C05125 | 2-(alpha-Hydroxyethyl)thiamine diphosphate |
| C15972 | Enzyme N6-(lipoyl)lysine |
| C15973 | Enzyme N6-(dihydrolipoyl)lysine |
| C16255 | [Dihydrolipoyllysine-residue acetyltransferase] S-acetyldihydrolipoyllysine |
|
|---|
| Reference |
|
|---|
| Authors |
Nishizuka Y, Seyama Y, Ikai A, Ishimura Y, Kawaguchi A (eds). |
|---|
| Title |
[Cellular Functions and Metabolic Maps] (In Japanese) |
|---|
| Journal |
Tokyo Kagaku Dojin (1997) |
|---|
Related
pathway |
| ko00010 | Glycolysis / Gluconeogenesis |
| ko00250 | Alanine, aspartate and glutamate metabolism |
| ko00260 | Glycine, serine and threonine metabolism |
| ko00290 | Valine, leucine and isoleucine biosynthesis |
| ko00522 | Biosynthesis of 12-, 14- and 16-membered macrolides |
| ko00630 | Glyoxylate and dicarboxylate metabolism |
| ko00760 | Nicotinate and nicotinamide metabolism |
| ko01056 | Biosynthesis of type II polyketide backbone |
| ko01059 | Biosynthesis of enediyne antibiotics |
|
|---|
|
|