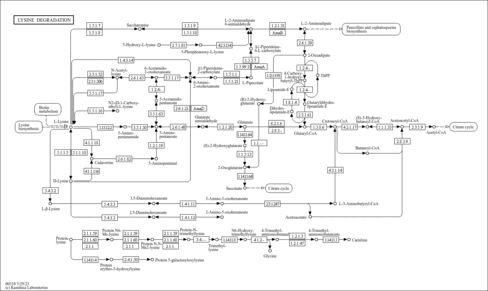
 | | PATHWAY: rn00310 | |
| Entry |
|
|---|
| Name |
Lysine degradation |
|---|
| Class |
Metabolism; Amino acid metabolism
|
|---|
| Pathway map |
|
|---|
| Module |
| M00032 | Lysine degradation, lysine => saccharopine => acetoacetyl-CoA [PATH:rn00310] |
| M00956 | Lysine degradation, bacteria, L-lysine => succinate [PATH:rn00310] |
| M00957 | Lysine degradation, bacteria, L-lysine => glutarate => succinate/acetyl-CoA [PATH:rn00310] |
| M00960 | Lysine degradation, bacteria, L-lysine => D-lysine => succinate [PATH:rn00310] |
|
|---|
| Other DBs |
|
|---|
| Reaction |
| R00238 | acetyl-CoA:acetyl-CoA C-acetyltransferase |
| R00447 | L-lysine:oxygen 2-oxidoreductase (deaminating) |
| R00449 | L-lysine:oxygen 2-oxidoreductase (decarboxylating) |
| R00452 | N2-(D-1-carboxylethyl)-L-lysine:NADP+ oxidoreductase (L-lysine-forming) |
| R00454 | acetyl-CoA:L-lysine N6-acetyltransferase |
| R00458 | N6-acetyl-L-lysine amidohydrolase |
| R00461 | L-lysine 2,3-aminomutase |
| R00462 | L-lysine carboxy-lyase (cadaverine-forming) |
| R00715 | N6-(L-1,3-dicarboxypropyl)-L-lysine:NAD+ oxidoreductase (L-lysine-forming) |
| R00716 | N6-(L-1,3-dicarboxypropyl)-L-lysine:NADP+ oxidoreductase (L-lysine-forming) |
| R01171 | butanoyl-CoA:NAD+ trans-2-oxidoreductase |
| R01365 | butanoyl-CoA:acetoacetate CoA-transferase |
| R01620 | acetyl-phosphate:L-lysine N6-acetyltransferase |
| R01939 | L-2-aminoadipate:2-oxoglutarate aminotransferase |
| R01975 | (S)-3-hydroxybutanoyl-CoA:NAD+ oxidoreductase |
| R02201 | L-pipecolate:NAD+ 2-oxidoreductase |
| R02203 | L-pipecolate:NADP+ 2-oxidoreductase |
| R02204 | L-pipecolate:oxygen 1,6-oxidoreductase |
| R02205 | L-pipecolate:(acceptor) 1,6-oxidoreductase |
| R02273 | 5-aminopentanamide amidohydrolase |
| R02274 | 5-aminopentanoate:2-oxoglutarate aminotransferase |
| R02276 | 5-acetamidopentanoate amidohydrolase |
| R02313 | N6-(L-1,3-dicarboxypropyl)-L-lysine:NAD+ oxidoreductase; N6-(L-1,3-dicarboxypropyl)-L-lysine:NAD+ oxidoreductase (L-glutamate-forming) |
| R02315 | N6-(L-1,3-dicarboxypropyl)-L-lysine:NADP+ oxidoreductase |
| R02317 | (S)-2-amino-6-oxohexanoate hydro-lyase (spontaneous) |
| R02397 | 4-trimethylammoniobutanoate,2-oxoglutarate:oxygen oxidoreductase (3-hydroxylating) |
| R02401 | glutarate-semialdehyde:NAD+ oxidoreductase |
| R02402 | glutarate:CoA ligase (ADP-forming) |
| R02488 | glutaryl-CoA:electron-transfer flavoprotein 2,3-oxidoreductase (decarboxylating) |
| R02571 | glutaryl-CoA:enzyme-N6-(dihydrolipoyl)lysine S-glutaryltransferase |
| R02851 | D-lysine:2-oxocarboxylate aminotransferase |
| R02852 | D-2,6-diaminohexanoic acid 5,6-aminomutase |
| R03026 | (S)-3-hydroxybutanoyl-CoA hydro-lyase |
| R03030 | L-3-aminobutyryl-CoA ammonia-lyase |
| R03102 | L-2-aminoadipate-6-semialdehyde:NAD+ 6-oxidoreductase |
| R03103 | L-2-aminoadipate-6-semialdehyde:NADP+ 6-oxidoreductase |
| R03275 | (3S)-3,6-diaminohexanoate 5,6-aminomutase |
| R03283 | 4-trimethylammoniobutanal:NAD+ 1-oxidoreductase; 4-trimethylammoniobutanal:NAD+ oxidoreductase |
| R03349 | L-erythro-3,5-diaminohexanoate:NAD+ oxidoreductase (deaminating) |
| R03376 | [procollagen]-L-lysine,2-oxoglutarate:oxygen oxidoreductase (5-hydroxylating) |
| R03378 | GTP:5-hydroxy-L-lysine O-phosphotransferase |
| R03380 | UDP-alpha-D-galactose:procollagen-5-hydroxy-L-lysine D-galactosyltransferase |
| R03451 | N6,N6,N6-trimethyl-L-lysine,2-oxoglutarate:oxygen oxidoreductase (3-hydroxylating) |
| R04029 | N6-acetyl-L-lysine:2-oxoglutarate aminotransferase |
| R04174 | 6-acetamido-2-oxohexanoate amidohydrolase |
| R04687 | 2,5-diaminohexanoate:NAD+ oxidoreductase (deaminating) |
| R04688 | 2,5-diaminohexanoate:NADP+ oxidoreductase (deaminating) |
| R04866 | S-adenosyl-L-methionine:cytochrome-c-L-lysine N6-methyltransferase |
| R04867 | S-adenosyl-L-methionine:cytochrome-c-L-lysine N6-methyltransferase |
| R07618 | enzyme N6-(dihydrolipoyl)lysine:NAD+ oxidoreductase |
| R09738 | butanoyl-CoA:NADP+ 2,3-oxidoreductase |
| R10270 | (5R)-5-phosphooxy-L-lysine phosphate-lyase (deaminating; (S)-2-amino-6-oxohexanoate-forming) |
| R10564 | (5S)-5-amino-3-oxohexanoate:acetyl-CoA ethylamine transferase |
| R12214 | cadaverine:2-oxoglutarate aminotransferase |
| R12215 | 5-aminopentanal:NAD+ 1-oxidoreductase |
| R12216 | glutarate,2-oxoglutarate:oxygen oxidoreductase ((S)-2-hydroxyglutarate-forming) |
| R12217 | (S)-2-hydroxyglutarate:quinone oxidoreductase |
| R13011 | 2-oxoadipate dioxygenase/carboxy lyase |
|
|---|
| Compound |
| C00449 | N6-(L-1,3-Dicarboxypropyl)-L-lysine |
| C00450 | (S)-2,3,4,5-Tetrahydropyridine-2-carboxylate |
| C01142 | (3S)-3,6-Diaminohexanoate |
| C01144 | (S)-3-Hydroxybutanoyl-CoA |
| C01149 | 4-Trimethylammoniobutanal |
| C01181 | 4-Trimethylammoniobutanoate |
| C01186 | (3S,5S)-3,5-Diaminohexanoate |
| C01211 | Procollagen 5-hydroxy-L-lysine |
| C01259 | (3S)-3-Hydroxy-N6,N6,N6-trimethyl-L-lysine |
| C03656 | (S)-5-Amino-3-oxohexanoic acid |
| C03793 | N6,N6,N6-Trimethyl-L-lysine |
| C04076 | L-2-Aminoadipate 6-semialdehyde |
| C04092 | Delta1-Piperideine-2-carboxylate |
| C04487 | 5-(D-Galactosyloxy)-L-lysine-procollagen |
| C05161 | (2R,5S)-2,5-Diaminohexanoate |
| C05544 | Protein N6-methyl-L-lysine |
| C05545 | Protein N6,N6-dimethyl-L-lysine |
| C05546 | Protein N6,N6,N6-trimethyl-L-lysine |
| C05548 | 6-Acetamido-2-oxohexanoate |
| C06157 | [Dihydrolipoyllysine-residue succinyltransferase] S-glutaryldihydrolipoyllysine |
| C15972 | Enzyme N6-(lipoyl)lysine |
| C15973 | Enzyme N6-(dihydrolipoyl)lysine |
| C22667 | 4-Carboxy-1-hydroxybutyryl-ThPP |
|
|---|
| Reference |
|
|---|
| Authors |
Vaz FM, Wanders RJ. |
|---|
| Title |
Carnitine biosynthesis in mammals. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Large PJ, Robertson A. |
|---|
| Title |
The route of lysine breakdown in Candida tropicalis. |
|---|
| Journal |
|
|---|
Related
pathway |
| rn00311 | Penicillin and cephalosporin biosynthesis |
|
|---|
| KO pathway |
|
|---|
|
|