 | | PATHWAY: paer00405 | |
| Entry |
|
|---|
| Name |
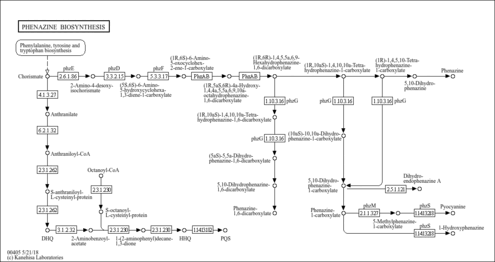
Phenazine biosynthesis - Pseudomonas aeruginosa PA1R |
|---|
| Class |
Metabolism; Biosynthesis of other secondary metabolites
|
|---|
| Pathway map |
|
|---|
| Module |
|
|---|
| Organism |
Pseudomonas aeruginosa PA1R [GN: paer]
|
|---|
| Gene |
|
|---|
| Compound |
| C11848 | 2-Heptyl-3-hydroxy-quinolone |
| C12119 | Phenazine-1,6-dicarboxylic acid |
| C18054 | 2-Amino-2-deoxyisochorismate |
| C19830 | (5S,6S)-6-Amino-5-hydroxycyclohexa-1,3-diene-1-carboxylate |
| C19831 | (1R,6S)-6-Amino-5-oxocyclohex-2-ene-1-carboxylate |
| C20643 | 2-Heptyl-4(1H)-quinolone |
| C20981 | 5,10-Dihydrophenazine-1-carboxylate |
| C20982 | 9-(Dimethylallyl)-5,10-dihydrophenazine-1-carboxylate |
| C21295 | 5-Methylphenazine-1-carboxylate |
| C21408 | (1R,6R)-1,4,5,5a,6,9-Hexahydrophenazine-1,6-dicarboxylate |
| C21409 | (1R,10aS)-1,4,10,10a-Tetrahydrophenazine-1,6-dicarboxylate |
| C21410 | (5aS)-5,5a-Dihydrophenazine-1,6-dicarboxylate |
| C21411 | (1R,10aS)-1,4,10,10a-Tetrahydrophenazine-1-carboxylate |
| C21412 | (10aS)-10,10a-Dihydrophenazine-1-carboxylate |
| C21413 | (1R)-1,4,5,10-Tetrahydrophenazine-1-carboxylate |
| C21442 | Phenazine-1-carboxylate |
| C21452 | (2-Aminobenzoyl)acetyl-CoA |
| C21453 | (2-Aminobenzoyl)acetate |
| C21473 | (1R,5aS,6R)-4a-Hydroxy-1,4,4a,5,5a,6,9,10a-octahydrophenazine-1,6-dicarboxylate |
| C21474 | 5,10-Dihydrophenazine-1,6-dicarboxylate |
| C21488 | S-Anthraniloyl-L-cysteinyl-protein |
| C21489 | S-Octanoyl-L-cysteinyl-protein |
| C21490 | 1-(2-Aminophenyl)decane-1,3-dione |
|
|---|
| Reference |
|
|---|
| Authors |
Xu N, Ahuja EG, Janning P, Mavrodi DV, Thomashow LS, Blankenfeldt W |
|---|
| Title |
Trapped intermediates in crystals of the FMN-dependent oxidase PhzG provide insight into the final steps of phenazine biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Pierson LS 3rd, Pierson EA |
|---|
| Title |
Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Ahuja EG, Janning P, Mentel M, Graebsch A, Breinbauer R, Hiller W, Costisella B, Thomashow LS, Mavrodi DV, Blankenfeldt W |
|---|
| Title |
PhzA/B catalyzes the formation of the tricycle in phenazine biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Mentel M, Ahuja EG, Mavrodi DV, Breinbauer R, Thomashow LS, Blankenfeldt W |
|---|
| Title |
Of two make one: the biosynthesis of phenazines. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Parsons JF, Calabrese K, Eisenstein E, Ladner JE |
|---|
| Title |
Structure of the phenazine biosynthesis enzyme PhzG. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS |
|---|
| Title |
Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. |
|---|
| Journal |
|
|---|
Related
pathway |
| paer00400 | Phenylalanine, tyrosine and tryptophan biosynthesis |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|
|
|