 | | PATHWAY: raq00620 | |
| Entry |
|
|---|
| Name |
Pyruvate metabolism - Rahnella aquatilis CIP 78.65 = ATCC 33071 |
|---|
| Class |
Metabolism; Carbohydrate metabolism
|
|---|
| Pathway map |
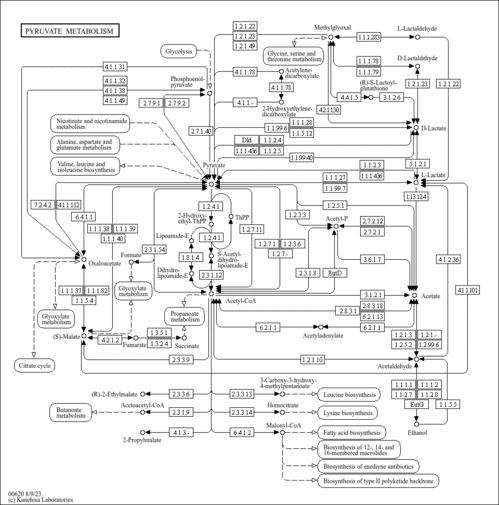
|
|---|
| Module |
| raq_M00579 | Phosphate acetyltransferase-acetate kinase pathway, acetyl-CoA => acetate [PATH:raq00620] |
|
|---|
| Other DBs |
|
|---|
| Organism |
Rahnella aquatilis CIP 78.65 = ATCC 33071 [GN: raq]
|
|---|
| Gene |
| Rahaq2_4904 | hydro-lyase, Fe-S type, tartrate/fumarate subfamily,hydro-lyase family enzyme, Fe-S type, tartrate/fumarate subfamily [KO:K01676] [EC:4.2.1.2] |
|
|---|
| Compound |
| C01251 | (R)-2-Hydroxybutane-1,2,4-tricarboxylate |
| C03451 | (R)-S-Lactoylglutathione |
| C03981 | 2-Hydroxyethylenedicarboxylate |
| C05125 | 2-(alpha-Hydroxyethyl)thiamine diphosphate |
| C15972 | Enzyme N6-(lipoyl)lysine |
| C15973 | Enzyme N6-(dihydrolipoyl)lysine |
| C16255 | [Dihydrolipoyllysine-residue acetyltransferase] S-acetyldihydrolipoyllysine |
|
|---|
| Reference |
|
|---|
| Authors |
Nishizuka Y, Seyama Y, Ikai A, Ishimura Y, Kawaguchi A (eds). |
|---|
| Title |
[Cellular Functions and Metabolic Maps] (In Japanese) |
|---|
| Journal |
Tokyo Kagaku Dojin (1997) |
|---|
Related
pathway |
| raq00250 | Alanine, aspartate and glutamate metabolism |
| raq00260 | Glycine, serine and threonine metabolism |
| raq00290 | Valine, leucine and isoleucine biosynthesis |
| raq00630 | Glyoxylate and dicarboxylate metabolism |
| raq00760 | Nicotinate and nicotinamide metabolism |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|
|
|