| Entry |
|
|---|
| Name |
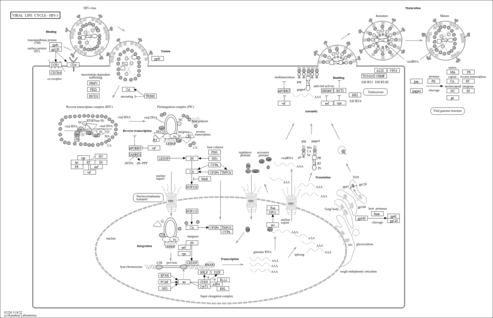
Viral life cycle - HIV-1 - Takifugu rubripes (torafugu) |
|---|
| Class |
Genetic Information Processing; Information processing in viruses
|
|---|
| Pathway map |
|
|---|
| Organism |
Takifugu rubripes (torafugu) [GN: tru]
|
|---|
| Gene |
| 101063041 | deoxynucleoside triphosphate triphosphohydrolase SAMHD1-like [KO:K22544] [EC:3.1.5.-] |
| 101070722 | deoxynucleoside triphosphate triphosphohydrolase SAMHD1-like [KO:K22544] [EC:3.1.5.-] |
| 105416260 | deoxynucleoside triphosphate triphosphohydrolase SAMHD1-like [KO:K22544] [EC:3.1.5.-] |
| 101070741 | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1-like [KO:K11648] |
| 101067729 | smarcb1; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 [KO:K11648] |
| 101070366 | cpsf7; cleavage and polyadenylation specificity factor subunit 7 isoform X1 [KO:K14398] |
| 101066960 | cpsf6; cleavage and polyadenylation specificity factor subunit 6 isoform X3 [KO:K14398] |
| 101077614 | E3 SUMO-protein ligase RanBP2 isoform X1 [KO:K12172] [EC:2.3.2.-] |
| 101078617 | nup153; nuclear pore complex protein Nup153 isoform X1 [KO:K14296] |
| 101072365 | RNA polymerase II elongation factor ELL2-like isoform X1 [KO:K15183] |
| 101075996 | nelfe; LOW QUALITY PROTEIN: negative elongation factor E [KO:K15182] |
| 101064061 | supt5h; transcription elongation factor SPT5 isoform X6 [KO:K15172] |
| 101075142 | hgs; hepatocyte growth factor-regulated tyrosine kinase substrate isoform X4 [KO:K12182] |
| 101079670 | tumor susceptibility gene 101 protein-like isoform X2 [KO:K12183] |
| 101071813 | pdcd6ip; programmed cell death 6-interacting protein isoform X2 [KO:K12200] |
| 101066795 | vps4a; vacuolar protein sorting-associated protein 4A [KO:K12196] |
| 101077130 | vacuolar protein sorting-associated protein 4B-like isoform X1 [KO:K12196] |
| 101069250 | vps4b; vacuolar protein sorting-associated protein 4B isoform X1 [KO:K12196] |
|
|---|
| Reference |
|
|---|
| Authors |
Li G, De Clercq E |
|---|
| Title |
HIV Genome-Wide Protein Associations: a Review of 30 Years of Research. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Tagliamonte M, Tornesello ML, Buonaguro FM, Buonaguro L |
|---|
| Title |
Conformational HIV-1 envelope on particulate structures: a tool for chemokine coreceptor binding studies. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Novikova M, Zhang Y, Freed EO, Peng K |
|---|
| Title |
Multiple Roles of HIV-1 Capsid during the Virus Replication Cycle. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Rawle DJ, Harrich D |
|---|
| Title |
Toward the "unravelling" of HIV: Host cell factors involved in HIV-1 core uncoating. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Le Sage V, Mouland AJ, Valiente-Echeverria F |
|---|
| Title |
Roles of HIV-1 capsid in viral replication and immune evasion. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Murphy RE, Saad JS |
|---|
| Title |
The Interplay between HIV-1 Gag Binding to the Plasma Membrane and Env Incorporation. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
He N, Zhou Q |
|---|
| Title |
New insights into the control of HIV-1 transcription: when Tat meets the 7SK snRNP and super elongation complex (SEC). |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Pincetic A, Leis J |
|---|
| Title |
The Mechanism of Budding of Retroviruses From Cell Membranes. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kleinpeter AB, Freed EO |
|---|
| Title |
HIV-1 Maturation: Lessons Learned from Inhibitors. |
|---|
| Journal |
|
|---|
Related
pathway |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|