| Entry |
|
|---|
| Name |
Ascorbate and aldarate metabolism - Arachis ipaensis |
|---|
| Class |
Metabolism; Carbohydrate metabolism
|
|---|
| Pathway map |
| aip00053 | Ascorbate and aldarate metabolism |
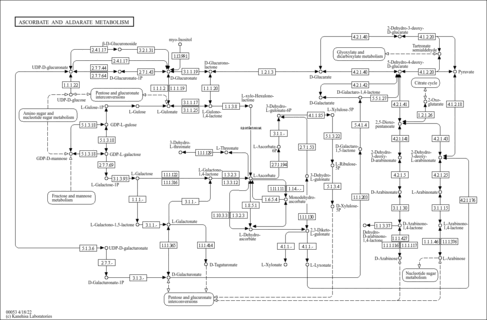
|
|---|
| Module |
|
|---|
| Other DBs |
|
|---|
| Organism |
Arachis ipaensis [GN: aip]
|
|---|
| Gene |
|
|---|
| Compound |
| C00652 | D-Arabinono-1,4-lactone |
| C00679 | 5-Dehydro-4-deoxy-D-glucarate |
| C00684 | 2-Dehydro-3-deoxy-L-arabinonate |
| C01114 | L-Arabinono-1,4-lactone |
| C01115 | L-Galactono-1,4-lactone |
| C01146 | 2-Hydroxy-3-oxopropanoate |
| C03826 | 2-Dehydro-3-deoxy-D-xylonate |
| C03921 | 2-Dehydro-3-deoxy-D-glucarate |
| C04037 | 1-Phospho-alpha-D-galacturonate |
| C04575 | (4R,5S)-4,5,6-Trihydroxy-2,3-dioxohexanoate |
| C05385 | D-Glucuronate 1-phosphate |
| C06316 | Dehydro-D-arabinono-1,4-lactone |
| C14899 | 3-Dehydro-L-gulonate 6-phosphate |
| C15926 | beta-L-Galactose 1-phosphate |
| C16186 | L-Ascorbate 6-phosphate |
| C20889 | D-Galactaro-1,5-lactone |
| C20896 | D-Galactaro-1,4-lactone |
| C21955 | L-Galactono-1,5-lactone |
|
|---|
| Reference |
|
|---|
| Authors |
Linster CL, Van Schaftingen E |
|---|
| Title |
Vitamin C. Biosynthesis, recycling and degradation in mammals. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Valpuesta V, Botella MA. |
|---|
| Title |
Biosynthesis of L-ascorbic acid in plants: new pathways for an old antioxidant. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Davey MW, Gilot C, Persiau G, Ostergaard J, Han Y, Bauw GC, Van Montagu MC. |
|---|
| Title |
Ascorbate biosynthesis in Arabidopsis cell suspension culture. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Conklin PL, Gatzek S, Wheeler GL, Dowdle J, Raymond MJ, Rolinski S, Isupov M, Littlechild JA, Smirnoff N. |
|---|
| Title |
Arabidopsis thaliana VTC4 encodes L-galactose-1-P phosphatase, a plant ascorbic acid biosynthetic enzyme. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Linster CL, Gomez TA, Christensen KC, Adler LN, Young BD, Brenner C, Clarke SG. |
|---|
| Title |
Arabidopsis VTC2 Encodes a GDP-L-Galactose Phosphorylase, the Last Unknown Enzyme in the Smirnoff-Wheeler Pathway to Ascorbic Acid in Plants. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Campos E, Baldoma L, Aguilar J, Badia J. |
|---|
| Title |
Regulation of expression of the divergent ulaG and ulaABCDEF operons involved in LaAscorbate dissimilation in Escherichia coli. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Zhang Z, Aboulwafa M, Smith MH, Saier MH Jr. |
|---|
| Title |
The ascorbate transporter of Escherichia coli. |
|---|
| Journal |
|
|---|
Related
pathway |
| aip00040 | Pentose and glucuronate interconversions |
| aip00051 | Fructose and mannose metabolism |
| aip00520 | Amino sugar and nucleotide sugar metabolism |
| aip00630 | Glyoxylate and dicarboxylate metabolism |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|