| Entry |
|
|---|
| Name |
Retinol metabolism - Haliotis rubra (blacklip abalone) |
|---|
| Class |
Metabolism; Metabolism of cofactors and vitamins
|
|---|
| Pathway map |
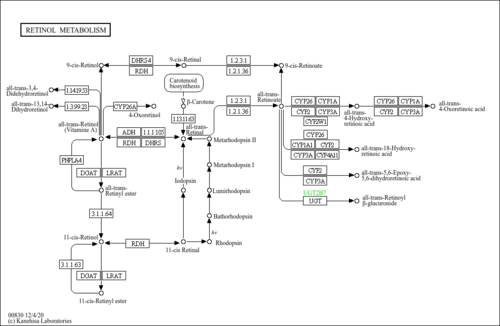
|
|---|
| Other DBs |
|
|---|
| Organism |
Haliotis rubra (blacklip abalone) [GN: hrj]
|
|---|
| Gene |
| 124254246 | dehydrogenase/reductase SDR family member 4-like isoform X1 [KO:K11147] [EC:1.1.-.-] |
| 124281125 | dehydrogenase/reductase SDR family member 9-like isoform X1 [KO:K11154] [EC:1.1.1.-] |
| 124262196 | patatin-like phospholipase domain-containing protein 4 [KO:K11157] [EC:3.1.1.-] |
|
|---|
| Compound |
| C03455 | 11-cis-Retinyl palmitate |
| C11061 | all-trans-Retinoyl-beta-glucuronide |
| C15492 | all-trans-13,14-Dihydroretinol |
| C16677 | all-trans-4-Hydroxyretinoic acid |
| C16678 | all-trans-4-Oxoretinoic acid |
| C16679 | all-trans-18-Hydroxyretinoic acid |
| C16680 | all-trans-5,6-Epoxyretinoic acid |
| C21797 | all-trans-3,4-Didehydroretinol |
|
|---|
| Reference |
|
|---|
| Authors |
Travis GH, Golczak M, Moise AR, Palczewski K. |
|---|
| Title |
Diseases caused by defects in the visual cycle: retinoids as potential therapeutic agents. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Muniz A, Villazana-Espinoza ET, Hatch AL, Trevino SG, Allen DM, Tsin AT. |
|---|
| Title |
A novel cone visual cycle in the cone-dominated retina. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Marill J, Cresteil T, Lanotte M, Chabot GG. |
|---|
| Title |
Identification of human cytochrome P450s involved in the formation of all-trans-retinoic acid principal metabolites. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Nezzar H, Chiambaretta F, Marceau G, Blanchon L, Faye B, Dechelotte P, Rigal D, Sapin V. |
|---|
| Title |
Molecular and metabolic retinoid pathways in the human ocular surface. |
|---|
| Journal |
Mol Vis 13:1641-50 (2007) |
|---|
| Reference |
|
|---|
| Authors |
Blomhoff R, Blomhoff HK. |
|---|
| Title |
Overview of retinoid metabolism and function. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Liden M, Eriksson U. |
|---|
| Title |
Understanding retinol metabolism: structure and function of retinol dehydrogenases. |
|---|
| Journal |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|