| Entry |
|
|---|
| Name |
Biosynthesis of various other secondary metabolites |
|---|
| Including |
Ditryptophenaline biosynthesis, Fumiquinazoline D biosynthesis, Paerucumarin biosynthesis, Staphyloferrin B biosynthesis, Cyclooctatin biosynthesis, Lovastatin biosynthesis, Grixazone biosynthesis, Staphyloferrin A biosynthesis, Ethynylserine biosynthesis, Aerobactin biosynthesis |
|---|
| Class |
Metabolism; Biosynthesis of other secondary metabolites
|
|---|
| Pathway map |
| rn00997 | Biosynthesis of various other secondary metabolites |
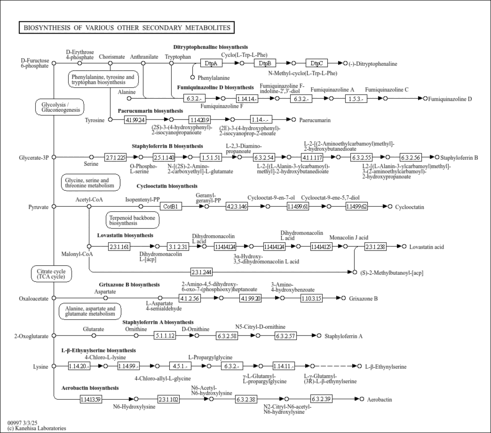
|
|---|
| Module |
| M00875 | Staphyloferrin B biosynthesis, L-serine => staphyloferrin B [PATH:rn00997] |
| M00876 | Staphyloferrin A biosynthesis, L-ornithine => staphyloferrin A [PATH:rn00997] |
| M00891 | Ditryptophenaline biosynthesis, tryptophan + phenylalanine => ditryptophenaline [PATH:rn00997] |
| M00893 | Lovastatin biosynthesis, malonyl-CoA => lovastatin acid [PATH:rn00997] |
| M00901 | Fumiquinazoline biosynthesis, tryptophan + alanine + anthranilate => fumiquinazoline [PATH:rn00997] |
| M00905 | Grixazone biosynthesis, aspartate 4-semialdehyde => grixazone B [PATH:rn00997] |
| M00906 | Ethynylserine biosynthesis, lysine => ethynylserine [PATH:rn00997] |
| M00918 | Aerobactin biosynthesis, lysine => aerobactin [PATH:rn00997] |
| M00921 | Cyclooctatin biosynthesis, dimethylallyl-PP + isopentenyl-PP => cyclooctatin [PATH:rn00997] |
|
|---|
| Reaction |
| R00448 | L-lysine,NADPH:oxygen oxidoreductase (6-hydroxylating) |
| R01658 | dimethylallyl-diphosphate:isopentenyl-diphosphate dimethylallyltranstransferase |
| R02003 | geranyl-diphosphate:isopentenyl-diphosphate geranyltrans-transferase |
| R02061 | trans,trans-farnesyl-diphosphate:isopentenyl-diphosphate farnesyltranstransferase |
| R03168 | acetyl-CoA:N6-hydroxy-L-lysine 6-acetyltransferase |
| R07251 | acyl-CoA:malonyl-CoA C-acyltransferase (dihydromonacolin L-[acp]-forming) |
| R10090 | citrate:N6-acetyl-N6-hydroxy-L-lysine ligase (AMP-forming) |
| R10091 | N2-citryl-N6-acetyl-N6-hydroxy-L-lysine:N6-acetyl-N6-hydroxy-L-lysine ligase (AMP-forming) |
| R10469 | 2-amino-4,5-dihydroxy-6-oxo-7-(phosphooxy)heptanoate hydro-lyase (cyclizing, 3-amino-4-hydroxybenzoate-forming) |
| R10673 | 2-amino-4,5-dihydroxy-6-oxo-7-(phosphooxy)heptanoate L-aspartate-4-semialdehyde-lyase (glycerone-phosphate-forming) |
| R10720 | 3-amino-4-hydroxybenzoate:N-acetyl-L-cysteine:oxygen oxidoreductase |
| R10763 | geranylgeranyl-diphosphate diphosphate-lyase (cyclooctat-9-en-7-ol-forming) |
| R10791 | dihydromonacolin L acid,[reduced NADPH---hemoprotein reductase]:oxygen oxidoreductase (3-hydroxylating) |
| R10792 | 3alpha-hydroxy-3,5-dihydromonacolin L acid hydro-lyase |
| R10794 | monacolin L acid,[reduced NADPH---hemoprotein reductase]:oxygen oxidoreductase (8-hydroxylating) |
| R11126 | monacolin-J-acid:(S)-2-methylbutanoyl-[acp] (S)-2-methylbutanoyltransferase |
| R11187 | dihydromonacolin L-[acyl-carrier protein] hydrolase |
| R11445 | malonyl-CoA:malonyl-CoA C-acyltransferase (decarboxylating, methylating, 2-methylbutanoate-forming) |
| R11655 | N-[(2S)-2-amino-2-carboxyethyl]-L-glutamate:NAD+ dehydrogenase (L-2,3-diaminopropanoate-forming) |
| R11705 | O-phospho-L-serine:L-glutamate N-(2S)-2-amino-2-carboxyethyltransferase |
| R12081 | (2S)-3-(4-hydroxyphenyl)-2-isocyanopropanoate,2-oxoglutarate:oxygen oxidoreductase |
| R12150 | L-tyrosine:D-ribulose-5-phosphate lyase (isonitrile-forming) |
| R12177 | cyclooctat-9-en-7-ol,hydrogen-donor:oxygen oxidoreductase (5-hydroxylating) |
| R12178 | cyclooctat-9-ene-5,7-diol,hydrogen-donor:oxygen oxidoreductase (18-hydroxylating) |
| R12308 | L-2,3-diaminopropanoate:citrate ligase (AMP, 2-[(L-alanin-3-ylcarbamoyl)methyl]-2-hydroxybutanedioate-forming) |
| R12309 | 2-[(L-alanin-3-ylcarbamoyl)methyl]-2-hydroxybutanedioate carboxy-lyase (2-[(2-aminoethylcarbamoyl)methyl]-2-hydroxybutanedioate-forming) |
| R12311 | 2-[(2-aminoethylcarbamoyl)methyl]-2-hydroxybutanedioate:L-2,3-diaminopropanoate ligase {2-[(L-alanin-3-ylcarbamoyl)methyl]-3-(2-aminoethylcarbamoyl)-2-hydroxypropanoate-forming} |
| R12312 | 2-[(L-alanin-3-ylcarbamoyl)methyl]-3-(2-aminoethylcarbamoyl)-2-hydroxypropanoate:2-oxoglutarate ligase (staphyloferrin B-forming) |
| R12344 | ATP:L-serine 3-phosphotransferase |
| R12353 | D-ornithine:citrate ligase (AMP-forming) |
| R12354 | N5-[(S)-citryl]-D-ornithine:citrate ligase (AMP, staphyloferrin A-forming) |
| R12407 | L-lysine,2-oxoglutarate:oxygen oxidoreductase (4-halogenating) |
| R12408 | 4-chloro-L-lysine,hydrogen-donor:oxygen oxidoreductase |
| R12411 | gamma-L-glutamyl-L-propargylglycine,2-oxoglutarate:oxygen oxidoreductase |
| R12414 | S-adenosyl-L-methionine:cyclo(L-Trp-L-Phe) N-methyltransferase |
|
|---|
| Compound |
| C00129 | Isopentenyl diphosphate |
| C00279 | D-Erythrose 4-phosphate |
| C00353 | Geranylgeranyl diphosphate |
| C00441 | L-Aspartate 4-semialdehyde |
| C03401 | L-2,3-Diaminopropanoate |
| C03955 | N6-Acetyl-N6-hydroxy-L-lysine |
| C04075 | 2-Amino-4-chloro-4-pentenoic acid |
| C12115 | 3-Amino-4-hydroxybenzoate |
| C20333 | N2-Citryl-N6-acetyl-N6-hydroxy-L-lysine |
| C20644 | 2-Amino-4,5-dihydroxy-6-oxo-7-(phosphooxy)heptanoate |
| C20851 | Dihydromonacolin L acid |
| C20852 | 3alpha-Hydroxy-3,5-dihydromonacolin L acid |
| C21108 | Dihydromonacolin L-[acyl-carrier protein] |
| C21129 | (S)-2-Methylbutanoyl-[acp] |
| C21559 | N-[(2S)-2-Amino-2-carboxyethyl]-L-glutamate |
| C21889 | (2S)-3-(4-Hydroxyphenyl)-2-isocyanopropanoate |
| C21891 | (2E)-3-(4-Hydroxyphenyl)-2-isocyanoprop-2-enoate |
| C21978 | Cyclooctat-9-ene-5,7-diol |
| C22072 | 2-[(L-Alanin-3-ylcarbamoyl)methyl]-2-hydroxybutanedioate |
| C22073 | 2-[(2-Aminoethylcarbamoyl)methyl]-2-hydroxybutanedioate |
| C22074 | 2-[(L-Alanin-3-ylcarbamoyl)methyl]-3-(2-aminoethylcarbamoyl)-2-hydroxypropanoate |
| C22139 | gamma-L-Glutamyl-L-propargylglycine |
| C22140 | L-gamma-Glutamyl-(3R)-L-beta-ethynylserine |
| C22143 | N-Methyl-cyclo(L-Trp-L-Phe) |
| C22146 | Fumiquinazoline F-indoline-2',3'-diol |
|
|---|
| Reference |
|
|---|
| Authors |
Saruwatari T, Yagishita F, Mino T, Noguchi H, Hotta K, Watanabe K |
|---|
| Title |
Cytochrome P450 as dimerization catalyst in diketopiperazine alkaloid biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Lim FY, Ames B, Walsh CT, Keller NP |
|---|
| Title |
Co-ordination between BrlA regulation and secretion of the oxidoreductase FmqD directs selective accumulation of fumiquinazoline C to conidial tissues in Aspergillus fumigatus. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Clarke-Pearson MF, Brady SF |
|---|
| Title |
Paerucumarin, a new metabolite produced by the pvc gene cluster from Pseudomonas aeruginosa. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Cheung J, Beasley FC, Liu S, Lajoie GA, Heinrichs DE |
|---|
| Title |
Molecular characterization of staphyloferrin B biosynthesis in Staphylococcus aureus. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kim SY, Zhao P, Igarashi M, Sawa R, Tomita T, Nishiyama M, Kuzuyama T |
|---|
| Title |
Cloning and heterologous expression of the cyclooctatin biosynthetic gene cluster afford a diterpene cyclase and two p450 hydroxylases. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Xu W, Chooi YH, Choi JW, Li S, Vederas JC, Da Silva NA, Tang Y |
|---|
| Title |
LovG: the thioesterase required for dihydromonacolin L release and lovastatin nonaketide synthase turnover in lovastatin biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Suzuki H, Ohnishi Y, Furusho Y, Sakuda S, Horinouchi S |
|---|
| Title |
Novel benzene ring biosynthesis from C(3) and C(4) primary metabolites by two enzymes. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Cotton JL, Tao J, Balibar CJ |
|---|
| Title |
Identification and characterization of the Staphylococcus aureus gene cluster coding for staphyloferrin A. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Marchand JA, Neugebauer ME, Ing MC, Lin CI, Pelton JG, Chang MCY |
|---|
| Title |
Discovery of a pathway for terminal-alkyne amino acid biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Neilands JB |
|---|
| Title |
Mechanism and regulation of synthesis of aerobactin in Escherichia coli K12 (pColV-K30). |
|---|
| Journal |
|
|---|
Related
pathway |
| rn00010 | Glycolysis / Gluconeogenesis |
| rn00250 | Alanine, aspartate and glutamate metabolism |
| rn00260 | Glycine, serine and threonine metabolism |
| rn00400 | Phenylalanine, tyrosine and tryptophan biosynthesis |
| rn00900 | Terpenoid backbone biosynthesis |
|
|---|
| KO pathway |
|
|---|