| Entry |
|
|---|
| Name |
Pentose phosphate pathway |
|---|
| Description |
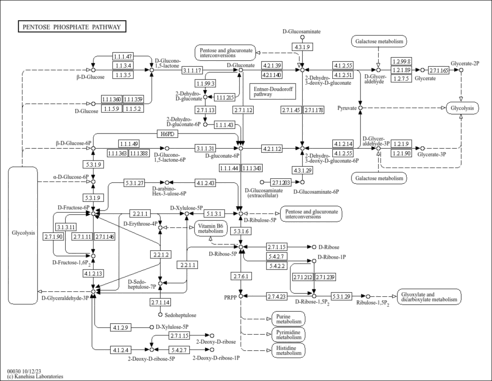
The pentose phosphate pathway is a process of glucose turnover that produces NADPH as reducing equivalents and pentoses as essential parts of nucleotides. There are two different phases in the pathway. One is irreversible oxidative phase in which glucose-6P is converted to ribulose-5P by oxidative decarboxylation, and NADPH is generated [MD: M00006]. The other is reversible non-oxidative phase in which phosphorylated sugars are interconverted to generate xylulose-5P, ribulose-5P, and ribose-5P [MD: M00007]. Phosphoribosyl pyrophosphate (PRPP) formed from ribose-5P [MD: M00005] is an activated compound used in the biosynthesis of histidine and purine/pyrimidine nucleotides. This pathway map also shows the Entner-Doudoroff pathway where 6-P-gluconate is dehydrated and then cleaved into pyruvate and glyceraldehyde-3P [MD: M00008].
|
|---|
| Class |
Metabolism; Carbohydrate metabolism
|
|---|
| Pathway map |
|
|---|
| Module |
| M00004 | Pentose phosphate pathway (Pentose phosphate cycle) [PATH:rn00030] |
| M00006 | Pentose phosphate pathway, oxidative phase, glucose 6P => ribulose 5P [PATH:rn00030] |
| M00007 | Pentose phosphate pathway, non-oxidative phase, fructose 6P => ribose 5P [PATH:rn00030] |
| M00008 | Entner-Doudoroff pathway, glucose-6P => glyceraldehyde-3P + pyruvate [PATH:rn00030] |
| M00061 | D-Glucuronate degradation, D-glucuronate => pyruvate + D-glyceraldehyde 3P [PATH:rn00030] |
| M00308 | Semi-phosphorylative Entner-Doudoroff pathway, gluconate => glycerate-3P [PATH:rn00030] |
| M00309 | Non-phosphorylative Entner-Doudoroff pathway, gluconate/galactonate => glycerate [PATH:rn00030] |
| M00345 | Formaldehyde assimilation, ribulose monophosphate pathway [PATH:rn00030] |
| M00580 | Pentose phosphate pathway, archaea, fructose 6P => ribose 5P [PATH:rn00030] |
| M00631 | D-Galacturonate degradation (bacteria), D-galacturonate => pyruvate + D-glyceraldehyde 3P [PATH:rn00030] |
| M00633 | Semi-phosphorylative Entner-Doudoroff pathway, gluconate/galactonate => glycerate-3P [PATH:rn00030] |
| M00968 | Pentose bisphosphate pathway (nucleoside degradation), archaea, nucleoside/NMP => 3-PGA/glycerone phosphate [PATH:rn00030] |
|
|---|
| Other DBs |
|
|---|
| Reaction |
| R00305 | D-glucose:quinone 1-oxidoreductase |
| R00756 | ATP:D-fructose-6-phosphate 1-phosphotransferase |
| R00762 | D-fructose-1,6-bisphosphate 1-phosphohydrolase |
| R00764 | diphosphate:D-fructose-6-phosphate 1-phosphotransferase |
| R01049 | ATP:D-ribose-5-phosphate diphosphotransferase |
| R01051 | ATP:D-ribose 5-phosphotransferase |
| R01056 | D-ribose-5-phosphate aldose-ketose-isomerase |
| R01057 | D-ribose 1,5-phosphomutase |
| R01058 | D-glyceraldehyde 3-phosphate:NADP+ oxidoreductase |
| R01066 | 2-deoxy-D-ribose-5-phosphate acetaldehyde-lyase (D-glyceraldehyde-3-phosphate-forming) |
| R01067 | D-fructose 6-phosphate:D-glyceraldehyde-3-phosphate glycolaldehyde transferase |
| R01068 | D-fructose-1,6-bisphosphate D-glyceraldehyde-3-phosphate-lyase (glycerone-phosphate-forming) |
| R01519 | D-glucono-1,5-lactone lactonohydrolase |
| R01520 | beta-D-glucose:NAD+ 1-oxoreductase |
| R01521 | beta-D-glucose:NADP+ 1-oxoreductase |
| R01522 | beta-D-glucose:oxygen 1-oxidoreductase |
| R01528 | 6-phospho-D-gluconate:NADP+ 2-oxidoreductase (decarboxylating) |
| R01529 | D-ribulose-5-phosphate 3-epimerase |
| R01538 | D-gluconate hydro-lyase (2-dehydro-3-deoxy-D-gluconate-forming) |
| R01541 | ATP:2-dehydro-3-deoxy-D-gluconate 6-phosphotransferase |
| R01544 | 2-amino-2-deoxy-D-gluconate ammonia-lyase (isomerizing; 2-dehydro-3-deoxy-D-gluconate-forming) |
| R01621 | D-xylulose 5-phosphate D-glyceraldehyde-3-phosphate-lyase (adding phosphate; acetyl-phosphate-forming) |
| R01641 | sedoheptulose-7-phosphate:D-glyceraldehyde-3-phosphate glycolaldehyde transferase |
| R01737 | ATP:D-gluconate 6-phosphotransferase |
| R01739 | D-gluconate:NADP+ 2-oxidoreductase |
| R01741 | D-gluconate:(acceptor) 2-oxidoreductase |
| R01844 | ATP:sedoheptulose 7-phosphate |
| R02032 | 6-phospho-2-dehydro-D-gluconate:NAD+ 2-oxidoreductase |
| R02034 | 6-phospho-D-gluconate:NADP+ 2-oxidoreductase |
| R02035 | 6-phospho-D-glucono-1,5-lactone lactonohydrolase |
| R02036 | 6-phospho-D-gluconate hydro-lyase (2-dehydro-3-deoxy-6-phospho-D-gluconate-forming) |
| R02658 | ATP:2-dehydro-D-gluconate 6-phosphate |
| R02736 | beta-D-glucose-6-phosphate:NADP+ 1-oxoreductase |
| R02739 | alpha-D-glucose 6-phosphate ketol-isomerase |
| R02749 | 2-deoxy-D-ribose 1-phosphate 1,5-phosphomutase |
| R02750 | ATP:2-deoxy-D-ribose 5-phosphotransferase |
| R05338 | D-arabino-hex-3-ulose-6-phosphate formaldehyde-lyase (D-ribulose-5-phosphate-forming) |
| R05339 | D-arabino-hex-3-ulose-6-phosphate isomerase |
| R05605 | 2-dehydro-3-deoxy-6-phospho-D-gluconate D-glyceraldehyde-3-phosphate-lyase (pyruvate-forming) |
| R05805 | ADP:D-fructose-6-phosphate 1-phosphotransferase |
| R06620 | D-glucose:ubiquinone oxidoreductase |
| R06836 | ATP:ribose-1,5-bisphosphate phosphotransferase |
| R07147 | D-glucose:NADP+ 1-oxidoreductase |
| R08570 | 2-dehydro-3-deoxy-D-gluconate D-glyceraldehyde-lyase (pyruvate-forming) |
| R08571 | glyceraldehyde ferredoxin oxidoreductase |
| R08572 | ATP:(R)-glycerate 2-phosphotransferase |
| R08575 | sedoheptulose-7-phosphate:D-glyceraldehyde-3-phosphate glyceronetransferase |
| R10221 | 6-phospho-D-gluconate:NAD+ 2-oxidoreductase (decarboxylating) |
| R10324 | D-glyceraldehyde:acceptor oxidoreductase (FAD-containing) |
| R10408 | D-glucosaminate-6-phosphate ammonia-lyase; 2-amino-2-deoxy-D-gluconate 6-phosphate ammonia-lyase (2-dehydro-3-deoxy-6-phospho-D-gluconate-forming) |
| R10555 | alpha-D-ribose 1,5-bisphosphate aldose-ketose-isomerase |
| R10615 | D-glyceraldehyde:NADP+ oxidoreductase |
| R10860 | D-glyceraldehyde-3-phosphate:NAD+ oxidoreductase |
| R10907 | beta-D-glucose-6-phosphate:NAD+ 1-oxidoreductase |
| R11535 | ADP:alpha-D-ribose-1-phosphate 5-phosphotransferase |
| R13089 | alpha-D-ribose-1-phosphate 5-kinase (ATP) |
| R13199 | alpha-D-glucose-6-phosphate aldose-ketose-isomerase |
|
|---|
| Compound |
| C00118 | D-Glyceraldehyde 3-phosphate |
| C00119 | 5-Phospho-alpha-D-ribose 1-diphosphate |
| C00204 | 2-Dehydro-3-deoxy-D-gluconate |
| C00279 | D-Erythrose 4-phosphate |
| C00354 | D-Fructose 1,6-bisphosphate |
| C00620 | alpha-D-Ribose 1-phosphate |
| C00668 | alpha-D-Glucose 6-phosphate |
| C00672 | 2-Deoxy-D-ribose 1-phosphate |
| C00673 | 2-Deoxy-D-ribose 5-phosphate |
| C01151 | D-Ribose 1,5-bisphosphate |
| C01172 | beta-D-Glucose 6-phosphate |
| C01182 | D-Ribulose 1,5-bisphosphate |
| C01218 | 6-Phospho-2-dehydro-D-gluconate |
| C01236 | D-Glucono-1,5-lactone 6-phosphate |
| C03752 | 2-Amino-2-deoxy-D-gluconate |
| C04442 | 2-Dehydro-3-deoxy-6-phospho-D-gluconate |
| C05382 | Sedoheptulose 7-phosphate |
| C06019 | D-arabino-Hex-3-ulose 6-phosphate |
| C20589 | D-Glucosaminate-6-phosphate |
|
|---|
| Reference |
|
|---|
| Authors |
Nishizuka Y (ed). |
|---|
| Title |
[Metabolic Maps] (In Japanese) |
|---|
| Journal |
Tokyo Kagaku Dojin (1980) |
|---|
| Reference |
|
|---|
| Authors |
Nishizuka Y, Seyama Y, Ikai A, Ishimura Y, Kawaguchi A (eds). |
|---|
| Title |
[Cellular Functions and Metabolic Maps] (In Japanese) |
|---|
| Journal |
Tokyo Kagaku Dojin (1997) |
|---|
| Reference |
|
|---|
| Authors |
Michal G. |
|---|
| Title |
Biochemical Pathways |
|---|
| Journal |
Wiley (1999) |
|---|
| Reference |
|
|---|
| Authors |
Hove-Jensen B, Rosenkrantz TJ, Haldimann A, Wanner BL. |
|---|
| Title |
Escherichia coli phnN, encoding ribose 1,5-bisphosphokinase activity (phosphoribosyl diphosphate forming): dual role in phosphonate degradation and NAD biosynthesis pathways. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Orita I, Sato T, Yurimoto H, Kato N, Atomi H, Imanaka T, Sakai Y |
|---|
| Title |
The ribulose monophosphate pathway substitutes for the missing pentose phosphate pathway in the archaeon Thermococcus kodakaraensis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kato N, Yurimoto H, Thauer RK |
|---|
| Title |
The physiological role of the ribulose monophosphate pathway in bacteria and archaea. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kouril T, Wieloch P, Reimann J, Wagner M, Zaparty M, Albers SV, Schomburg D, Ruoff P, Siebers B |
|---|
| Title |
Unraveling the function of the two Entner-Doudoroff branches in the thermoacidophilic Crenarchaeon Sulfolobus solfataricus P2. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Reher M, Schonheit P |
|---|
| Title |
Glyceraldehyde dehydrogenases from the thermoacidophilic euryarchaeota Picrophilus torridus and Thermoplasma acidophilum, key enzymes of the non-phosphorylative Entner-Doudoroff pathway, constitute a novel enzyme family within the aldehyde dehydrogenase superfamily. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Reher M, Fuhrer T, Bott M, Schonheit P |
|---|
| Title |
The nonphosphorylative Entner-Doudoroff pathway in the thermoacidophilic euryarchaeon Picrophilus torridus involves a novel 2-keto-3-deoxygluconate- specific aldolase. |
|---|
| Journal |
|
|---|
Related
pathway |
| rn00010 | Glycolysis / Gluconeogenesis |
| rn00040 | Pentose and glucuronate interconversions |
| rn00630 | Glyoxylate and dicarboxylate metabolism |
|
|---|
| KO pathway |
|
|---|