| Entry |
|
|---|
| Name |
Pentose and glucuronate interconversions |
|---|
| Class |
Metabolism; Carbohydrate metabolism
|
|---|
| Pathway map |
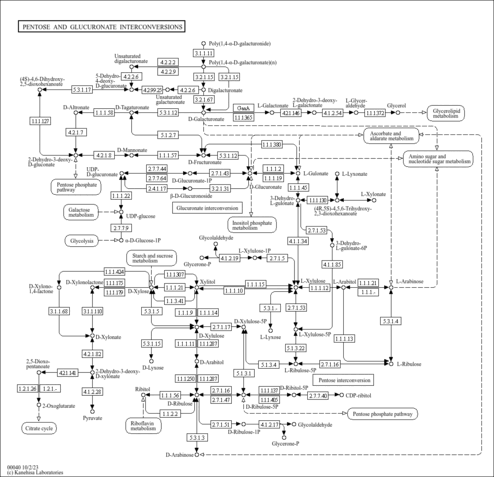
| rn00040 | Pentose and glucuronate interconversions |
|
|---|
| Module |
| M00061 | D-Glucuronate degradation, D-glucuronate => pyruvate + D-glyceraldehyde-3P [PATH:rn00040] |
| M00129 | Ascorbate biosynthesis, animals, glucose-1P => ascorbate [PATH:rn00040] |
| M00630 | D-Galacturonate degradation (fungi), D-galacturonate => glycerol [PATH:rn00040] |
| M00631 | D-Galacturonate degradation (bacteria), D-galacturonate => pyruvate + D-glyceraldehyde-3P [PATH:rn00040] |
| M00994 | UDP-GlcA biosynthesis, myo-inositol => GlcA => UDP-GlcA [PATH:rn00040] |
|
|---|
| Reaction |
| R00264 | 2,5-dioxopentanoate:NADP+ 5-oxidoreductase |
| R00286 | UDP-glucose:NAD+ 6-oxidoreductase |
| R00289 | UTP:alpha-D-glucose-1-phosphate uridylyltransferase |
| R01381 | UTP:1-phospho-alpha-D-glucuronate uridylyltransferase |
| R01383 | UDPglucuronate beta-D-glucuronosyltransferase (acceptor-unspecific) |
| R01429 | D-xylose:NAD+ 1-oxidoreductase |
| R01430 | D-xylose:NADP+ 1-oxidoreductase |
| R01431 | xylitol:NADP+ oxidoreductase |
| R01432 | D-xylose aldose-ketose-isomerase |
| R01476 | ATP:D-glucuronate 1-phosphotransferase |
| R01478 | beta-D-glucuronoside glucuronosohydrolase |
| R01481 | L-gulonate:NADP+ 6-oxidoreductase |
| R01482 | D-glucuronate aldose-ketose-isomerase |
| R01524 | D-ribitol-5-phosphate:NAD+ 2-oxidoreductase |
| R01525 | D-ribitol-5-phosphate:NADP+ 2-oxidoreductase |
| R01526 | ATP:D-ribulose 5-phosphotransferase |
| R01529 | D-ribulose-5-phosphate 3-epimerase |
| R01540 | D-altronate hydro-lyase (2-dehydro-3-deoxy-D-gluconate-forming) |
| R01542 | 2-dehydro-3-deoxy-D-gluconate:NAD+ 5-oxidoreductase |
| R01577 | D-arabinose aldose-ketose-isomerase |
| R01639 | ATP:D-xylulose 5-phosphotransferase |
| R01758 | L-arabitol:NAD+ 1-oxidoreductase |
| R01759 | L-arabitol:NADP+ 1-oxidoreductase |
| R01761 | L-arabinose aldose-ketose-isomerase |
| R01782 | 2-dehydro-3-deoxy-D-pentonate glycolaldehyde-lyase (pyruvate-forming) |
| R01785 | L-xylulose 1-phosphate glycolaldehyde-lyase (glycerone-phosphate-forming) |
| R01892 | ribitol:ferricytochrome-c 2-oxidoreductase |
| R01895 | ribitol:NAD+ 2-oxidoreductase |
| R01896 | xylitol:NAD+ 2-oxidoreductase (D-xylulose-forming) |
| R01898 | D-lyxose aldose-ketose-isomerase |
| R01901 | ATP:L-xylulose 5-phosphotransferase |
| R01902 | ATP:L-xylulose 1-phosphotransferase |
| R01903 | L-arabinitol:NAD+ 4-oxidoreductase (L-xylulose-forming) |
| R01904 | xylitol:NADP+ 4-oxidoreductase (L-xylulose-forming) |
| R01905 | 3-dehydro-L-gulonate carboxy-lyase |
| R01906 | L-lyxose aldose-ketose-isomerase |
| R01982 | poly(1,4-alpha-D-galacturonate) galacturonohydrolase |
| R01983 | D-galacturonate aldose-ketose-isomerase |
| R02360 | poly(1,4-alpha-D-galacturonate) glycanohydrolase |
| R02361 | (1->4)-alpha-D-galacturonan reducing-end-disaccharide-lyase |
| R02427 | D-xylono-1,5-lactone lactonohydrolase |
| R02428 | D-xylono-1,4-lactone lactonohydrolase |
| R02439 | ATP:L-ribulose 5-phosphotransferase |
| R02441 | L-arabitol:NAD+ 2-oxidoreductase (L-ribulose-forming) |
| R02454 | D-mannonate:NAD+ 5-oxidoreductase |
| R02555 | D-altronate:NAD+ 3-oxidoreductase |
| R02637 | 3-dehydro-L-gulonate:NAD+ 2-oxidoreductase |
| R02639 | 3-dehydro-L-gulonate:NADP+ 2-oxidoreductase |
| R02640 | L-gulonate:NAD+ 3-oxidoreductase |
| R02921 | CTP:D-ribitol-5-phosphate cytidylyltransferase |
| R03244 | L-ribulose-5-phosphate 3-epimerase; L-xylulose 5-phosphate 3-epimerase |
| R04382 | 4-(4-deoxy-alpha-D-galact-4-enuronosyl)-D-galacturonate lyase |
| R04383 | 5-dehydro-4-deoxy-D-glucuronate ketol-isomerase |
| R05604 | D-arabinitol:NAD 4-oxidoreductase |
| R05606 | D-mannonate hydro-lyase |
| R05831 | xylitol:NAD oxidoreductase |
| R05850 | L-ribulose-5-phosphate 4-epimerase |
| R07125 | 3-dehydro-L-gulonate-6-phosphate carboxy-lyase |
| R07127 | ATP:3-dehydro-L-gulonate 6-phosphotransferase |
| R07134 | D-arabinitol:NAD+ 2-oxidoreductase (D-ribulose-forming) |
| R07143 | D-arabinitol:NADP+ oxidoreductase |
| R07144 | D-arabinitol:NADP+ oxidoreductase |
| R07152 | xylitol:oxygen oxidoreductase |
| R07676 | L-galactonate:NADP+ oxidoreductase |
| R09186 | 2-dehydro-3-deoxy-D-arabinonate hydro-lyase (2,5-dioxopentanoate-forming) |
| R09477 | xylitol:NAD+ oxidoreductase |
| R10532 | L-galactonate hydro-lyase (2-dehydro-3-deoxy-L-galactonate-forming) |
| R10550 | 2-dehydro-3-deoxy-L-galactonate L-glyceraldehyde-lyase (pyruvate-forming) |
| R10563 | glycerol:NADP+ oxidoreductase (D/L-glyceraldehyde-forming) |
| R10565 | L-galactonate:NAD+ oxidoreductase |
| R10848 | L-gulonate:NAD+ 5-oxidoreductase |
| R10861 | 3,6-anhydro-alpha-L-galactopyranose:NAD+ 1-oxidoredutase |
| R10862 | 3,6-anhydro-alpha-L-galactopyranose:NADP+ 1-oxidoredutase |
| R10900 | 3,6-anhydro-L-galactonate lyase (ring-opening) |
| R11035 | 2-dehydro-3-deoxy-L-galactonate:NAD+ 5-oxidoreductase |
| R11624 | D-tagaturonate 3-epimerase |
| R11768 | 2,5-dioxopentanoate:NAD+ 5-oxidoreductase |
| R12643 | D-xylose:NADP+ 1-oxidoreductase (D-xylono-1,4-lactone-forming) |
| R12695 | ATP:D-ribulose 1-phosphotransferase |
| R12696 | D-ribulose 1-phosphate glycolaldehyde-lyase (glycerone-phosphate-forming) |
| R13196 | 4,5-unsaturated pyranuronate lyase (ring-opening) |
|
|---|
| Compound |
| C00204 | 2-Dehydro-3-deoxy-D-gluconate |
| C03826 | 2-Dehydro-3-deoxy-D-xylonate |
| C04053 | 5-Dehydro-4-deoxy-D-glucuronate |
| C04349 | (4S)-4,6-Dihydroxy-2,5-dioxohexanoate |
| C04575 | (4R,5S)-4,5,6-Trihydroxy-2,3-dioxohexanoate |
| C05385 | D-Glucuronate 1-phosphate |
| C06118 | 4-(4-Deoxy-alpha-D-gluc-4-enuronosyl)-D-galacturonate |
| C14899 | 3-Dehydro-L-gulonate 6-phosphate |
| C20680 | 2-Dehydro-3-deoxy-L-galactonate |
| C20902 | 3,6-Anhydro-alpha-L-galactopyranose |
| C20903 | 3,6-Anhydro-L-galactonate |
| C22712 | 4-Deoxy-L-threo-hex-4-enopyranuronate |
|
|---|
| Reference |
|
|---|
| Authors |
Yew WS, Gerlt JA. |
|---|
| Title |
Utilization of L-ascorbate by Escherichia coli K-12: assignments of functions to products of the yjf-sga and yia-sgb operons. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Yew WS, Akana J, Wise EL, Rayment I, Gerlt JA |
|---|
| Title |
Evolution of enzymatic activities in the orotidine 5'-monophosphate decarboxylase suprafamily: enhancing the promiscuous D-arabino-hex-3-ulose 6-phosphate synthase reaction catalyzed by 3-keto-L-gulonate 6-phosphate decarboxylase. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Yasueda H, Kawahara Y, Sugimoto S. |
|---|
| Title |
Bacillus subtilis yckG and yckF encode two key enzymes of the ribulose monophosphate pathway used by methylotrophs, and yckH is required for their expression. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Ibanez E, Gimenez R, Pedraza T, Baldoma L, Aguilar J, Badia J |
|---|
| Title |
Role of the yiaR and yiaS genes of Escherichia coli in metabolism of endogenously formed L-xylulose. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Orita I, Yurimoto H, Hirai R, Kawarabayasi Y, Sakai Y, Kato N |
|---|
| Title |
The archaeon Pyrococcus horikoshii possesses a bifunctional enzyme for formaldehyde fixation via the ribulose monophosphate pathway. |
|---|
| Journal |
|
|---|
Related
pathway |
| rn00010 | Glycolysis / Gluconeogenesis |
| rn00053 | Ascorbate and aldarate metabolism |
| rn00520 | Amino sugar and nucleotide sugar metabolism |
| rn00562 | Inositol phosphate metabolism |
|
|---|
| KO pathway |
|
|---|