| Entry |
|
|---|
| Name |
Arginine biosynthesis - Oryza sativa japonica (Japanese rice) |
|---|
| Class |
Metabolism; Amino acid metabolism
|
|---|
| Pathway map |
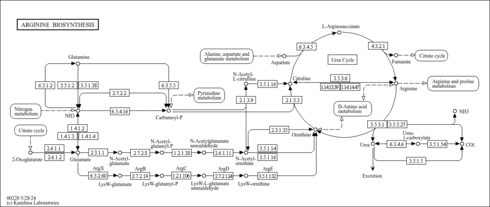
|
|---|
| Module |
|
|---|
| Other DBs |
|
|---|
| Organism |
Oryza sativa japonica (Japanese rice) [GN: osa]
|
|---|
| Gene |
| 4326346 | CARB; carbamoyl phosphate synthase arginine-specific large chain, chloroplastic [KO:K01955] [EC:6.3.5.5] |
| 4330474 | 49D11.14; carbamoyl phosphate synthase small chain, chloroplastic isoform 1 [KO:K01956] [EC:6.3.5.5] |
| 4333458 | AGPR; probable N-acetyl-gamma-glutamyl-phosphate reductase, chloroplastic [KO:K00145] [EC:1.2.1.38] |
|
|---|
| Compound |
| C01250 | N-Acetyl-L-glutamate 5-semialdehyde |
| C03406 | N-(L-Arginino)succinate |
| C04133 | N-Acetyl-L-glutamate 5-phosphate |
| C20949 | LysW-L-glutamyl 5-phosphate |
| C20950 | LysW-L-glutamate 5-semialdehyde |
|
|---|
Related
pathway |
| osa00250 | Alanine, aspartate and glutamate metabolism |
| osa00330 | Arginine and proline metabolism |
|
|---|
| KO pathway |
|
|---|