| Entry |
|
|---|
| Name |
3-hydroxypropionate:NADP+ oxidoreductase
|
|---|
| Definition |
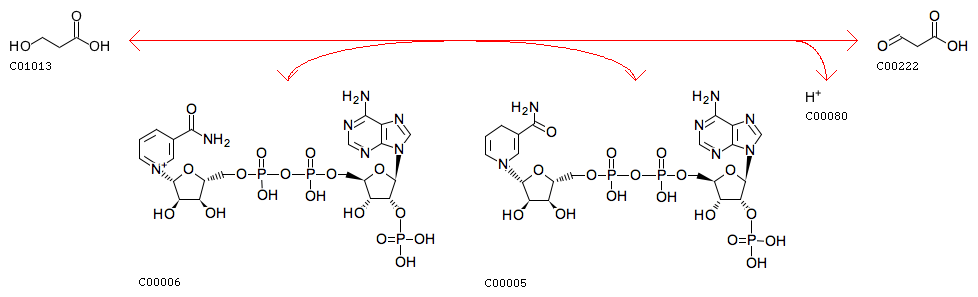
3-Hydroxypropanoate + NADP+ <=> 3-Oxopropanoate + NADPH + H+
|
|---|
| Equation |
|
|---|
|
 |
|---|
| Reaction class |
|
|---|
| Enzyme |
|
|---|
| Pathway |
| rn00720 | Other carbon fixation pathways |
| rn01120 | Microbial metabolism in diverse environments |
|
|---|
| Module |
| M00375 | Hydroxypropionate-hydroxybutylate cycle |
| M00376 | 3-Hydroxypropionate bi-cycle |
| M00939 | Pyrimidine degradation, uracil => 3-hydroxypropanoate |
|
|---|
| Brite |
Enzymatic reactions [BR:br08201]
1. Oxidoreductase reactions
1.1 Acting on the CH-OH group of donors
1.1.1 With NAD+ or NADP+ as acceptor
1.1.1.298
R09289 3-Hydroxypropanoate + NADP+ <=> 3-Oxopropanoate + NADPH + H+
1.1.1.-
R09289 3-Hydroxypropanoate + NADP+ <=> 3-Oxopropanoate + NADPH + H+
Overall reaction [br08210.html]
Carbon fixation
R09289
|
|---|
| Orthology |
| K09019 | 3-hydroxypropanoate dehydrogenase [EC:1.1.1.-] |
| K16066 | 3-hydroxy acid dehydrogenase / malonic semialdehyde reductase [EC:1.1.1.381 1.1.1.-] |
| K18602 | malonic semialdehyde reductase [EC:1.1.1.-] |
|
|---|
| LinkDB |
|
|---|