| Entry |
|
|---|
| Name |
Amino sugar and nucleotide sugar metabolism |
|---|
| Class |
Metabolism; Glycan biosynthesis and metabolism
|
|---|
| Pathway map |
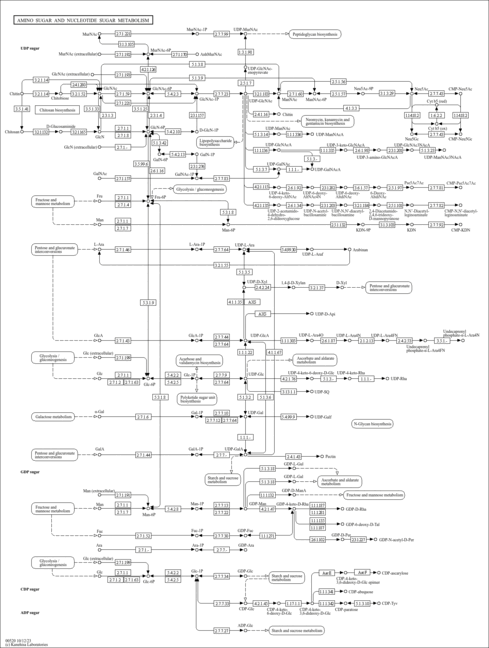
| rn00520 | Amino sugar and nucleotide sugar metabolism |
|
|---|
| Module |
| M00892 | UDP-GlcNAc biosynthesis, eukaryotes, Fru-6P => UDP-GlcNAc [PATH:rn00520] |
| M00909 | UDP-GlcNAc biosynthesis, prokaryotes, Fru-6P => UDP-GlcNAc [PATH:rn00520] |
| M00994 | UDP-GlcA biosynthesis, myo-inositol => GlcA => UDP-GlcA [PATH:rn00520] |
| M00995 | UDP-MurNAc biosynthesis, Fru-6P => UDP-MurNAc [PATH:rn00520] |
| M00996 | UDP-MurNAc biosynthesis, anhMurNAc => UDP-MurNAc [PATH:rn00520] |
| M00997 | UDP-Xyl/L-Ara biosynthesis, UDP-Glc => UDP-Xyl => UDP-L-Ala [PATH:rn00520] |
| M00999 | UDP-GlcA/GalA biosynthesis, UDP-Glc => UDP-GlcA => UDP-GalA [PATH:rn00520] |
| M01002 | UDP-GalNAc biosynthesis, Fru-6P => UDP-GalNAc [PATH:rn00520] |
| M01003 | UDP-GlcNAc biosynthesis, GlcNAc => UDP-GlcNAc [PATH:rn00520] |
| M01006 | GDP-D-Rha4N biosynthesis, Fru-6P => GDP-D-Rha4N [PATH:rn00520] |
|
|---|
| Other DBs |
|
|---|
| Reaction |
| R00022 | chitobiose N-acetylglucosaminohydrolase |
| R00286 | UDP-glucose:NAD+ 6-oxidoreductase |
| R00289 | UTP:alpha-D-glucose-1-phosphate uridylyltransferase |
| R00291 | UDP-glucose 4-epimerase; UDP-alpha-D-glucose 4-epimerase |
| R00416 | UTP:N-acetyl-alpha-D-glucosamine-1-phosphate uridylyltransferase |
| R00418 | UDP-N-acetyl-D-glucosamine 4-epimerase |
| R00420 | UDP-N-acetyl-D-glucosamine 2-epimerase |
| R00501 | UDP-alpha-D-galactose:NAD+ 6-oxidoreductase |
| R00502 | UTP:alpha-D-galactose-1-phosphate uridylyltransferase |
| R00505 | UDP-alpha-D-galactopyranose furanomutase |
| R00660 | phosphoenolpyruvate:UDP-N-acetyl-D-glucosamine 1-carboxyvinyl-transferase |
| R00760 | ATP:D-fructose 6-phosphotransferase |
| R00765 | D-glucosamine-6-phosphate aminohydrolase (ketol isomerizing); D-glucosamine-6-phosphate ketol-isomerase(deaminating) |
| R00768 | L-glutamine:D-fructose-6-phosphate isomerase (deaminating); L-glutamine:D-fructose-6-phosphate aminotransferase (hexose isomerizing) |
| R00772 | D-mannose-6-phosphate aldose-ketose-isomerase |
| R00880 | GDP-D-mannose:NAD+ 6-oxidoreductase |
| R00883 | GDP:D-mannose-1-phosphate guanylyltransferase |
| R00885 | GTP:alpha-D-mannose-1-phosphate guanylyltransferase |
| R00889 | GDP-mannose 3,5-epimerase |
| R00948 | ATP:alpha-D-glucose-1-phosphate adenylyltransferase |
| R00954 | GTP:alpha-D-glucose-1-phosphate guanylyltransferase |
| R00955 | UDP-glucose:alpha-D-galactose-1-phosphate uridylyltransferase |
| R00956 | CTP:alpha-D-glucose-1-phosphate cytidylyltransferase |
| R00959 | alpha-D-glucose 1-phosphate 1,6-phosphomutase |
| R01092 | ATP:alpha-D-galactose 1-phosphotransferase |
| R01200 | N-acetyl-D-glucosamine amidohydrolase |
| R01201 | ATP:N-acetyl-D-glucosamine 6-phosphotransferase |
| R01204 | acetyl-CoA:D-glucosamine N-acetyltransferase |
| R01206 | [1,4-(N-acetyl-beta-D-glucosaminyl)]n glycanohydrolase |
| R01207 | N-acetyl-D-glucosamine 2-epimerase |
| R01326 | ATP:D-mannose 6-phosphotransferase |
| R01381 | UTP:1-phospho-alpha-D-glucuronate uridylyltransferase |
| R01384 | UDP-D-glucuronate carboxy-lyase (UDP-D-xylose-forming) |
| R01385 | UDP-glucuronate 4-epimerase |
| R01386 | UDP-D-glucuronate carboxy-lyase (UDP-D-apiose-forming) |
| R01471 | UTP:alpha-D-xylose-1-phosphate uridylyltransferase |
| R01473 | UDP-L-arabinose 4-epimerase |
| R01476 | ATP:D-glucuronate 1-phosphotransferase |
| R01754 | ATP:L-arabinose 1-phosphotransferase |
| R01762 | alpha-L-arabinan arabinofuranohydrolase |
| R01786 | ATP:alpha-D-glucose 6-phosphotransferase |
| R01818 | D-mannose 6-phosphate 1,6-phosphomutase |
| R01951 | GTP:beta-L-fucose-1-phosphate guanylyltransferase |
| R01961 | ATP:D-glucosamine 6-phosphotransferase |
| R01966 | beta-D-glucosaminide glucosaminohydrolase |
| R01980 | ATP:D-galacturonate 1-phosphotransferase |
| R02058 | acetyl-CoA:D-glucosamine-6-phosphate N-acetyltransferase |
| R02059 | N-acetyl-D-glucosamine-6-phosphate amidohydrolase |
| R02060 | D-glucosamine 1-phosphate 1,6-phosphomutase; D-glucosamine 1,6-phosphomutase |
| R02087 | N-acetyl-D-glucosamine 6-phosphate 2-epimerase |
| R02189 | polyphosphate:D-glucose 6-phosphotransferase |
| R02328 | dTTP:alpha-D-glucose-1-phosphate thymidylyltransferase |
| R02334 | [1,4-(N-acetyl-beta-D-glucosaminyl)]n glycanohydrolase |
| R02335 | UDP-N-acetyl-D-glucosamine:chitin 4-beta-N-acetylglucosaminyl-transferase |
| R02630 | protein-N(pi)-phosphohistidine:D-mannose 6-phosphotransferase |
| R02631 | protein-N(pi)-phosphohistidine:D-glucosamine 6-phosphotransferase |
| R02634 | UTP:1-phospho-alpha-D-galacturonate uridylyltransferase; UDP-D-galacturonate pyrophospholylase |
| R02636 | UDP-D-galacturonate carboxy-lyase |
| R02705 | ATP:N-acyl-D-mannosamine 6-phosphotransferase |
| R02738 | protein-N(pi)-phosphohistidine:D-glucose 6-phosphotransferase |
| R02833 | chitosan N-acetylglucosaminohydrolase |
| R03077 | UTP:beta-L-arabinose-1-phosphate uridylyltransferase |
| R03161 | ATP:L-fucose 1-phosphotransferase |
| R03191 | UDP-N-acetylmuramate:NADP+ oxidoreductase |
| R03192 | UDP-N-acetylmuramate:NADP+ oxidoreductase |
| R03928 | UDP-D-xylose:1,4-beta-D-xylan 4-beta-D-xylosyltransferase |
| R05168 | N-acetyl-D-galactosamine 6-phosphate amidohydrolase |
| R05191 | UDP-D-galacturonate:1,4-alpha-poly-D-galacturonate 4-alpha-D-galacturonosyltransferase |
| R05199 | protein-N(pi)-phosphohistidine:N-acetyl-D-glucosamine 6-phosphotransferase |
| R05332 | acetyl-CoA:D-glucosamine-1-phosphate N-acetyltransferase |
| R05775 | UDP-6-sulfo-6-deoxyglucose sulfohydrolase |
| R07672 | GDP-mannose 5-epimerase |
| R08193 | N-acetyl-D-glucosamine 1-phosphate 1,6-phosphomutase |
| R08366 | protein-N(pi)-phosphohistidine:N-acetyl-D-galactosamine 6-phosphotransferase |
| R08367 | protein-N(pi)-phosphohistidine:D-galactosamine 6-phosphotransferase |
| R08555 | (R)-lactate hydro-lyase (adding N-acetyl-D-glucosamine 6-phosphate; N-acetylmuramate 6-phosphate-forming) |
| R08559 | protein-N(pi)-phosphohistidine:N-acetylmuramate 6-phosphotransferase |
| R08967 | ATP:N-acetyl-D-galactosamine 1-phosphotransferase |
| R08968 | ATP:N-acetyl-D-glucosamine 1-phosphotransferase |
| R09009 | UDP-L-arabinopyranose furanomutase |
| R09942 | N,N'-diacetylchitobiose:phosphate N-acetyl-D-glucosaminyltransferase |
| R10183 | UTP:N-acetyl-alpha-D-galactosamine-1-phosphate uridylyltransferase |
| R11024 | ATP:N-acetyl-D-muramate 1-phosphotransferase |
| R11025 | UTP:N-acetyl-alpha-D-muramate-1-phosphate uridylyltransferase |
| R11785 | N-acetyl-D-muramate 6-phosphate phosphohydrolase |
| R12220 | D-glucosamine 6-phosphate 4-epimerase |
| R12221 | alpha-D-galactosamine 1,6-phosphomutase |
| R12242 | acetyl-CoA:alpha-D-galactosamine-1-phosphate N-acetyltransferase |
| R12755 | ATP:1,6-anhydro-N-acetyl-beta-muramate 6-phosphotransferase |
| R12824 | ATP:D-arabinose 1-phosphotransferase |
| R12825 | GTP:alpha-D-arabinopyranose-1-phosphate guanylyltransferase |
| R13199 | alpha-D-glucose-6-phosphate aldose-ketose-isomerase |
|
|---|
| Compound |
| C00043 | UDP-N-acetyl-alpha-D-glucosamine |
| C00203 | UDP-N-acetyl-D-galactosamine |
| C00352 | D-Glucosamine 6-phosphate |
| C00357 | N-Acetyl-D-glucosamine 6-phosphate |
| C00446 | alpha-D-Galactose 1-phosphate |
| C00668 | alpha-D-Glucose 6-phosphate |
| C01132 | N-Acetyl-D-galactosamine |
| C01170 | UDP-N-acetyl-D-mannosamine |
| C03733 | UDP-alpha-D-galactofuranose |
| C03737 | alpha-D-Xylose 1-phosphate |
| C03906 | beta-L-Arabinose 1-phosphate |
| C04037 | 1-Phospho-alpha-D-galacturonate |
| C04257 | N-Acetyl-D-mannosamine 6-phosphate |
| C04501 | N-Acetyl-alpha-D-glucosamine 1-phosphate |
| C04631 | UDP-N-acetyl-3-(1-carboxyvinyl)-D-glucosamine |
| C05385 | D-Glucuronate 1-phosphate |
| C06156 | alpha-D-Glucosamine 1-phosphate |
| C06376 | N-Acetyl-D-galactosamine 6-phosphate |
| C06377 | D-Galactosamine 6-phosphate |
| C16183 | alpha-D-Arabinopyranose 1-phosphate |
| C16698 | N-Acetylmuramic acid 6-phosphate |
| C18060 | N-Acetyl-alpha-D-galactosamine 1-phosphate |
| C19769 | 1,6-Anhydro-N-acetyl-beta-muramate |
| C21027 | N-Acetylmuramic acid alpha-1-phosphate |
| C22029 | alpha-D-Galactosamine 1-phosphate |
|
|---|
| Reference |
|
|---|
| Authors |
Wyk P, Reeves P |
|---|
| Title |
Identification and sequence of the gene for abequose synthase, which confers antigenic specificity on group B salmonellae: homology with galactose epimerase. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Thorson JS, Lo SF, Ploux O, He X, Liu HW |
|---|
| Title |
Studies of the biosynthesis of 3,6-dideoxyhexoses: molecular cloning and characterization of the asc (ascarylose) region from Yersinia pseudotuberculosis serogroup VA. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kessler AC, Haase A, Reeves PR |
|---|
| Title |
Molecular analysis of the 3,6-dideoxyhexose pathway genes of Yersinia pseudotuberculosis serogroup IIA. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Danese PN, Oliver GR, Barr K, Bowman GD, Rick PD, Silhavy TJ |
|---|
| Title |
Accumulation of the enterobacterial common antigen lipid II biosynthetic intermediate stimulates degP transcription in Escherichia coli. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Skurnik M, Peippo A, Ervela E |
|---|
| Title |
Characterization of the O-antigen gene clusters of Yersinia pseudotuberculosis and the cryptic O-antigen gene cluster of Yersinia pestis shows that the plague bacillus is most closely related to and has evolved from Y. pseudotuberculosis serotype O:1b. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, Garber RL, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody LL, Coulter SN, Folger KR, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong GK, Wu Z, Paulsen IT, Reizer J, Saier MH, Hancock RE, Lory S, Olson MV |
|---|
| Title |
Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Ringenberg M, Lichtensteiger C, Vimr E |
|---|
| Title |
Redirection of sialic acid metabolism in genetically engineered Escherichia coli. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
McCoy AJ, Sandlin RC, Maurelli AT |
|---|
| Title |
In vitro and in vivo functional activity of Chlamydia MurA, a UDP-N-acetylglucosamine enolpyruvyl transferase involved in peptidoglycan synthesis and fosfomycin resistance. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
JACOBSON B, DAVIDSON EA |
|---|
| Title |
Biosynthesis of uronic acids by skin enzymes. I. Uridine diphosphate-D-glucuronic acid-5-epimerase. |
|---|
| Journal |
J Biol Chem 237:638-42 (1962) |
|---|
| Reference |
|
|---|
| Authors |
Seifert GJ |
|---|
| Title |
Nucleotide sugar interconversions and cell wall biosynthesis: how to bring the inside to the outside. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Uehara T, Park JT |
|---|
| Title |
The N-acetyl-D-glucosamine kinase of Escherichia coli and its role in murein recycling. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Breazeale SD, Ribeiro AA, McClerren AL, Raetz CR. |
|---|
| Title |
A formyltransferase required for polymyxin resistance in Escherichia coli and the modification of lipid A with 4-Amino-4-deoxy-L-arabinose. Identification and function oF UDP-4-deoxy-4-formamido-L-arabinose. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Williams GJ, Breazeale SD, Raetz CR, Naismith JH. |
|---|
| Title |
Structure and function of both domains of ArnA, a dual function decarboxylase and a formyltransferase, involved in 4-amino-4-deoxy-L-arabinose biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Maliekal P, Vertommen D, Delpierre G, Van Schaftingen E |
|---|
| Title |
Identification of the sequence encoding N-acetylneuraminate-9-phosphate phosphatase. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Campbell CT, Yarema KJ |
|---|
| Title |
Large-scale approaches for glycobiology. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Uehara T, Suefuji K, Jaeger T, Mayer C, Park JT |
|---|
| Title |
MurQ Etherase is required by Escherichia coli in order to metabolize anhydro-N-acetylmuramic acid obtained either from the environment or from its own cell wall. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Lanot A, Hodge D, Jackson RG, George GL, Elias L, Lim EK, Vaistij FE, Bowles DJ |
|---|
| Title |
The glucosyltransferase UGT72E2 is responsible for monolignol 4-O-glucoside production in Arabidopsis thaliana. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Muller R, Morant M, Jarmer H, Nilsson L, Nielsen TH |
|---|
| Title |
Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Oka T, Nemoto T, Jigami Y |
|---|
| Title |
Functional analysis of Arabidopsis thaliana RHM2/MUM4, a multidomain protein involved in UDP-D-glucose to UDP-L-rhamnose conversion. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Greenberg DE, Porcella SF, Zelazny AM, Virtaneva K, Sturdevant DE, Kupko JJ 3rd, Barbian KD, Babar A, Dorward DW, Holland SM. |
|---|
| Title |
Genome sequence analysis of the emerging human pathogenic acetic acid bacterium Granulibacter bethesdensis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kotake T, Hojo S, Tajima N, Matsuoka K, Koyama T, Tsumuraya Y |
|---|
| Title |
A bifunctional enzyme with L-fucokinase and GDP-L-fucose pyrophosphorylase activities salvages free L-fucose in Arabidopsis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Vorholter FJ, Schneiker S, Goesmann A, Krause L, Bekel T, Kaiser O, Linke B, Patschkowski T, Ruckert C, Schmid J, Sidhu VK, Sieber V, Tauch A, Watt SA, Weisshaar B, Becker A, Niehaus K, Puhler A. |
|---|
| Title |
The genome of Xanthomonas campestris pv. campestris B100 and its use for the reconstruction of metabolic pathways involved in xanthan biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Albermann C, Piepersberg W |
|---|
| Title |
Expression and identification of the RfbE protein from Vibrio cholerae O1 and its use for the enzymatic synthesis of GDP-D-perosamine. |
|---|
| Journal |
|
|---|
Related
pathway |
| rn00010 | Glycolysis / Gluconeogenesis |
| rn00040 | Pentose and glucuronate interconversions |
| rn00051 | Fructose and mannose metabolism |
| rn00053 | Ascorbate and aldarate metabolism |
| rn00500 | Starch and sucrose metabolism |
| rn00523 | Polyketide sugar unit biosynthesis |
| rn00524 | Neomycin, kanamycin and gentamicin biosynthesis |
| rn00525 | Acarbose and validamycin biosynthesis |
| rn00540 | Lipopolysaccharide biosynthesis |
| rn00541 | Biosynthesis of various nucleotide sugars |
|
|---|
| KO pathway |
|
|---|