 | | PATHWAY: rn00620 | |
| Entry |
|
|---|
| Name |
Pyruvate metabolism |
|---|
| Class |
Metabolism; Carbohydrate metabolism
|
|---|
| Pathway map |
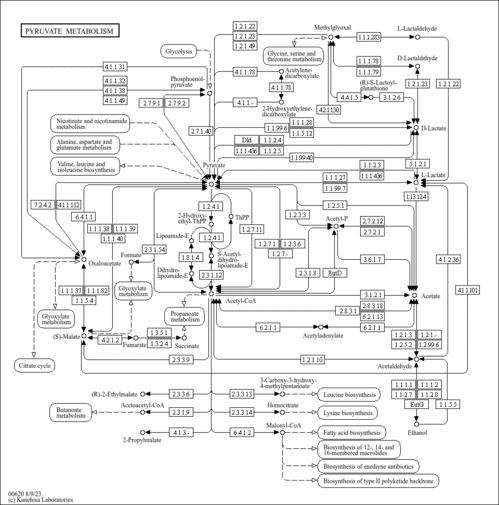
|
|---|
| Module |
| M00172 | C4-dicarboxylic acid cycle, NADP - malic enzyme type [PATH:rn00620] |
| M00579 | Phosphate acetyltransferase-acetate kinase pathway, acetyl-CoA => acetate [PATH:rn00620] |
|
|---|
| Other DBs |
|
|---|
| Reaction |
| R00014 | pyruvate:thiamin diphosphate acetaldehydetransferase (decarboxylating) |
| R00196 | (S)-lactate:ferricytochrome-c 2-oxidoreductase |
| R00197 | (R)-lactate:ferricytochrome-c 2-oxidoreductase |
| R00198 | (R)-lactate:ferricytochrome-c-553 2-oxidoreductase |
| R00199 | ATP:pyruvate,water phosphotransferase |
| R00200 | ATP:pyruvate 2-O-phosphotransferase |
| R00203 | pyruvaldehyde:NAD+ oxidoreductase |
| R00205 | 2-oxopropanal:NADP+ oxidoreductase |
| R00206 | ATP:pyruvate,phosphate phosphotransferase |
| R00207 | pyruvate:oxygen 2-oxidoreductase (phosphorylating) |
| R00211 | pyruvate:oxygen 2-oxidoreductase (CoA-acetylating) |
| R00212 | acetyl-CoA:formate C-acetyltransferase |
| R00214 | (S)-malate:NAD+ oxidoreductase (decarboxylating) |
| R00216 | (S)-malate:NADP+ oxidoreductase(oxaloacetate-decarboxylating) |
| R00217 | oxaloacetate carboxy-lyase (pyruvate-forming) |
| R00218 | acetylenedicarboxylate carboxy-lyase (pyruvate-forming) |
| R00219 | 2-hydroxyethylenedicarboxylate carboxy-lyase (pyruvate-forming) |
| R00228 | acetaldehyde:NAD+ oxidoreductase (CoA-acetylating) |
| R00229 | acetate:CoA ligase (ADP-forming) |
| R00230 | acetyl-CoA:phosphate acetyltransferase |
| R00236 | acetyl adenylate:CoA acetyltransferase |
| R00238 | acetyl-CoA:acetyl-CoA C-acetyltransferase |
| R00271 | acetyl-CoA:2-oxoglutarate C-acetyltransferase (thioester-hydrolysing, carboxymethyl forming) |
| R00297 | (R)-2-hydroxy-acid:acceptor 2-oxidoreductase |
| R00315 | ATP:acetate phosphotransferase |
| R00316 | ATP:acetate adenylyltransferase |
| R00317 | acetyl phosphate phosphohydrolase |
| R00319 | (S)-lactate:oxygen 2-oxidoreductase (decarboxylating) |
| R00320 | diphosphate:acetate phosphotransferase |
| R00326 | acetaldehyde:acceptor oxidoreductase |
| R00341 | ATP:oxaloacetate carboxy-lyase (transphosphorylating;phosphoenolpyruvate-forming) |
| R00342 | (S)-malate:NAD+ oxidoreductase |
| R00343 | (S)-malate:NADP+ oxidoreductase |
| R00344 | pyruvate:carbon-dioxide ligase (ADP-forming) |
| R00345 | phosphate:oxaloacetate carboxy-lyase (adding phosphate;phosphoenolpyruvate-forming) |
| R00346 | diphosphate:oxaloacetate carboxy-lyase (transphosphorylating;phosphoenolpyruvate-forming) |
| R00431 | GTP:oxaloacetate carboxy-lyase (adding GTP;phosphoenolpyruvate-forming) |
| R00472 | acetyl-CoA:glyoxylate C-acetyltransferase (thioester-hydrolysing, carboxymethyl-forming) |
| R00703 | (S)-lactate:NAD+ oxidoreductase |
| R00704 | (R)-lactate:NAD+ oxidoreductase |
| R00710 | acetaldehyde:NAD+ oxidoreductase |
| R00711 | acetaldehyde:NADP+ oxidoreductase |
| R00726 | ITP:oxaloacetate carboxy-lyase (adding ITP; phosphoenolpyruvate-forming) |
| R00742 | acetyl-CoA:carbon-dioxide ligase (ADP-forming) |
| R00746 | ethanol:NADP+ oxidoreductase |
| R00753 | (S)-lactate acetaldehyde-lyase (formate-forming) |
| R00754 | ethanol:NAD+ oxidoreductase |
| R00928 | acetyl-CoA:propanoate CoA-transferase |
| R00998 | acetyl-CoA:2-oxobutanoate C-acetyltransferase (thioester-hydrolysing, carboxymethyl-forming) |
| R01019 | acetaldehyde:pyrroloquinoline-quinone oxidoreductase |
| R01082 | (S)-malate hydro-lyase (fumarate-forming) |
| R01196 | pyruvate:ferredoxin 2-oxidoreductase (CoA-acetylating) |
| R01213 | acetyl-CoA:3-methyl-2-oxobutanoate C-acetyltransferase (thioester-hydrolysing, carboxymethyl-forming) |
| R01257 | (S)-malate:FAD oxidoreductase |
| R01446 | (S)-lactaldehyde:NAD+ oxidoreductase |
| R01447 | (S)-lactate:oxaloacetate oxidoreductase |
| R01735 | (R)-lactaldehyde:NAD+ 2-oxidoreductase |
| R01736 | (R)-S-lactoylglutathione hydrolase |
| R02164 | succinate:quinone oxidoreductase |
| R02260 | (S)-lactaldehyde:NADP+ oxidoreductase |
| R02320 | NTP:pyruvate O2-phosphotransferase |
| R02527 | (R)-lactaldehyde:NAD+ oxidoreductase |
| R02530 | (R)-S-lactoylglutathione methylglyoxal-lyase (isomerizing) |
| R02569 | acetyl-CoA:enzyme N6-(dihydrolipoyl)lysine S-acetyltransferase |
| R03145 | pyruvate:ubiquinone oxidoreductase |
| R04177 | 2-hydroxyethylenedicarboxylate hydro-lyase |
| R05198 | ethanol:cytochrome c oxidoreductase |
| R07618 | enzyme N6-(dihydrolipoyl)lysine:NAD+ oxidoreductase |
| R09127 | ethanol:cytochrome c oxidoreductase |
| R09479 | ethanol:quinone oxidoreductase |
| R09796 | (R)-lactate hydro-lyase |
| R10343 | succinyl-CoA:acetate CoA-transferase |
| R10866 | pyruvate:flavodoxin 2-oxidoreductase (CoA-acetylating) |
| R11074 | (S)-malate carboxy-lyase |
| R11591 | (R)-lactate:quinone 2-oxidoreductase |
| R11593 | (R)-2-hydroxyglutarate:pyruvate oxidoreductase [(R)-lactate-forming] |
| R12212 | oxaloacetate carboxy-lyase (pyruvate-forming; Na+-extruding) |
| R13049 | (R)-lactate:NAD+,ferredoxin oxidoreductase |
| R13050 | (S)-lactate:NAD+,ferredoxin oxidoreductase |
| R13137 | succinate:ferricytochrome-c oxidoreductase |
|
|---|
| Compound |
| C01251 | (R)-2-Hydroxybutane-1,2,4-tricarboxylate |
| C03451 | (R)-S-Lactoylglutathione |
| C03981 | 2-Hydroxyethylenedicarboxylate |
| C05125 | 2-(alpha-Hydroxyethyl)thiamine diphosphate |
| C15972 | Enzyme N6-(lipoyl)lysine |
| C15973 | Enzyme N6-(dihydrolipoyl)lysine |
| C16255 | [Dihydrolipoyllysine-residue acetyltransferase] S-acetyldihydrolipoyllysine |
|
|---|
| Reference |
|
|---|
| Authors |
Nishizuka Y, Seyama Y, Ikai A, Ishimura Y, Kawaguchi A (eds). |
|---|
| Title |
[Cellular Functions and Metabolic Maps] (In Japanese) |
|---|
| Journal |
Tokyo Kagaku Dojin (1997) |
|---|
Related
pathway |
| rn00010 | Glycolysis / Gluconeogenesis |
| rn00250 | Alanine, aspartate and glutamate metabolism |
| rn00260 | Glycine, serine and threonine metabolism |
| rn00290 | Valine, leucine and isoleucine biosynthesis |
| rn00522 | Biosynthesis of 12-, 14- and 16-membered macrolides |
| rn00630 | Glyoxylate and dicarboxylate metabolism |
| rn00760 | Nicotinate and nicotinamide metabolism |
| rn01056 | Biosynthesis of type II polyketide backbone |
| rn01059 | Biosynthesis of enediyne antibiotics |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|
|
|