 | | PATHWAY: xma00970 | |
| Entry |
|
|---|
| Name |
Aminoacyl-tRNA biosynthesis - Xiphophorus maculatus (southern platyfish) |
|---|
| Class |
Genetic Information Processing; Translation
|
|---|
| Pathway map |
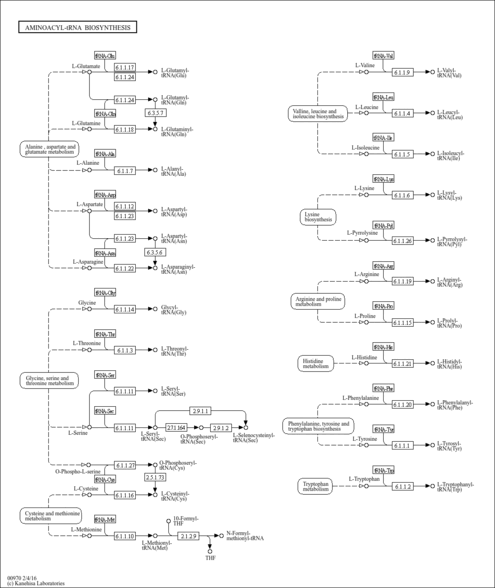
|
|---|
| Organism |
Xiphophorus maculatus (southern platyfish) [GN: xma]
|
|---|
| Gene |
|
|---|
| Compound |
| C00234 | 10-Formyltetrahydrofolate |
| C03402 | L-Asparaginyl-tRNA(Asn) |
| C03511 | L-Phenylalanyl-tRNA(Phe) |
| C03512 | L-Tryptophanyl-tRNA(Trp) |
| C06482 | L-Selenocysteinyl-tRNA(Sec) |
| C16638 | O-Phosphoseryl-tRNA(Sec) |
| C17022 | O-Phosphoseryl-tRNA(Cys) |
|
|---|
| Reference |
|
|---|
| Authors |
Sheppard K, Yuan J, Hohn MJ, Jester B, Devine KM, Soll D |
|---|
| Title |
From one amino acid to another: tRNA-dependent amino acid biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Polycarpo C, Ambrogelly A, Berube A, Winbush SM, McCloskey JA, Crain PF, Wood JL, Soll D |
|---|
| Title |
An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. |
|---|
| Journal |
|
|---|
Related
pathway |
| xma00250 | Alanine, aspartate and glutamate metabolism |
| xma00260 | Glycine, serine and threonine metabolism |
| xma00270 | Cysteine and methionine metabolism |
| xma00290 | Valine, leucine and isoleucine biosynthesis |
| xma00330 | Arginine and proline metabolism |
| xma00400 | Phenylalanine, tyrosine and tryptophan biosynthesis |
|
|---|
| KO pathway |
|
|---|
|
|