| Entry |
|
|---|
| Name |
Neomycin, kanamycin and gentamicin biosynthesis |
|---|
| Class |
Metabolism; Biosynthesis of other secondary metabolites
|
|---|
| Pathway map |
| rn00524 | Neomycin, kanamycin and gentamicin biosynthesis |
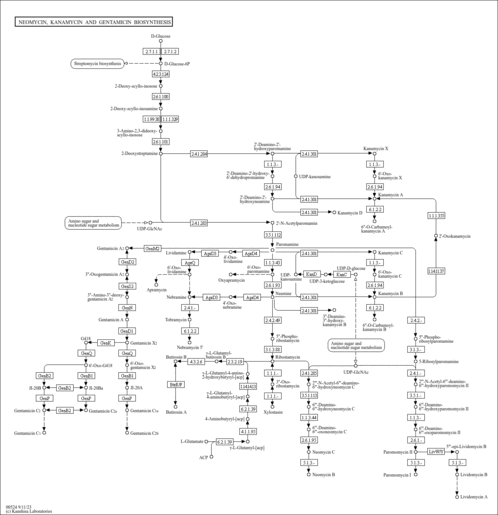
|
|---|
| Drug |
|
|---|
| Other DBs |
|
|---|
| Reaction |
| R00299 | ATP:D-glucose 6-phosphotransferase |
| R06586 | UDP-3-ketoglucose:L-glutamate aminotransferase |
| R06594 | UDP-glucose:NAD+ 3-oxidoreductase |
| R08617 | D-glucose-6-phosphate phosphate-lyase (2-deoxy-scyllo-inosose-forming) |
| R08890 | L-glutamine:2-deoxy-scyllo-inosose aminotransferase |
| R08891 | L-glutamine:3-amino-2,3-dideoxy-scyllo-inosose aminotransferase |
| R08892 | 2-deoxy-scyllo-inosamine:NAD+ oxidoreductase |
| R08893 | 2-deoxy-scyllo-inosamine:S-adenosyl-L-methionine 1-oxidoreductase |
| R08894 | UDP-N-acetyl-alpha-D-glucosamine:2-deoxystreptamine N-acetyl-D-glucosaminyltransferase |
| R08895 | 2'-N-acetylparomamine hydrolase (acetate-forming) |
| R08896 | paromamine:oxygen 6'-oxidoreductase |
| R08897 | neamine:2-oxoglutarate aminotransferase |
| R08898 | 5-phospho-alpha-D-ribose 1-diphosphate:neamine phosphoribosyltransferase; 5''-phosphoribostamycin:diphosphate phospho-alpha-D-ribosyltransferase |
| R08899 | 5''-phosphoribostamycin phosphohydrolase |
| R08900 | UDP-N-acetyl-alpha-D-glucosamine:ribostamycin N-acetyl-D-glucosaminyltransferase |
| R08901 | 2'''-acetyl-6'''-hydroxyneomycin-C hydrolase (acetate-forming) |
| R08902 | 6'''-deamino-6'''-hydroxyneomycin-C:oxygen 6'''-oxidoreductase |
| R08903 | 2-oxoglutarate:neomycin-C aminotransferase |
| R08904 | neomycin-C 5'''-epimerase |
| R08905 | L-glutamate:[acyl-carrier protein] ligase (ADP-forming) |
| R08906 | L-glutamate:4-aminobutyryl-[acp] ligase (ADP-forming) |
| R08907 | gamma-L-glutamyl-[acp] carboxy-lyase (4-aminobutyryl-[acp]-forming) |
| R08908 | 4-(gamma-L-glutamylamino)butanoyl-[acp],reduced-FMN:oxygen oxidoreductase |
| R08909 | 4-(gamma-L-glutamylamino)-(S)-2-hydroxybutanoyl-[acp]:ribostamycin 4-(gamma-L-glutamylamino)-(S)-2-hydroxybutanoate transferase |
| R08910 | gamma-L-glutamyl-butirosin-B gamma-glutamyl cyclotransferase (5-oxoproline-forming) |
| R10094 | 2'-deamino-2'-hydroxyneamine:2-oxoglutarate aminotransferase |
| R10096 | UDP-alpha-D-glucose:2-deoxystreptamine 4-O-alpha-D-glucosyltransferase |
| R10130 | 2-deoxy-scyllo-inosamine:NADP+ 1-oxidoreductase |
| R10207 | 2'-deamino-2'-hydroxyparomamine:oxygen 6'-oxidoreductase |
| R10312 | kanamycin-B,2-oxoglutarate:oxygen oxidoreductase (deaminating, 2'-hydroxylating) |
| R10417 | kanamycin-A:NADP+ oxidoreductase |
| R10424 | UDP-alpha-D-kanosamine:2'-deamino-2'-hydroxyneamine 1-alpha-D-kanosaminyltransferase |
| R10425 | UDP-alpha-D-kanosamine:neamine 1-alpha-D-kanosaminyltransferase |
| R10426 | UDP-alpha-D-kanosamine:paromamine 1-alpha-D-kanosaminyltransferase |
| R10427 | UDP-alpha-D-kanosamine:2'-deamino-2'-hydroxyparomamine 1-alpha-D-kanosaminyltransferase |
| R10767 | tobramycin:carbamoyl-phosphate ligase (AMP,phosphate-forming) |
| R10768 | kanamycin-A:carbamoyl-phosphate ligase (AMP,phosphate-forming) |
| R11265 | ribostamycin:NAD+ 3''-oxidoreductase |
| R11266 | xylostasin:NADP+ 3''-oxidoreductase |
| R11267 | kanamycin-X:oxygen 6'-oxidoreductase |
| R11268 | kanamycin-C:oxygen 6'-oxidoreductase |
| R11269 | kanamycin-A:2-oxoglutarate aminotransferase |
| R11270 | kanamycin-B:2-oxoglutarate aminotransferase |
| R11271 | UDP-alpha-D-glucose:2'-deamino-2'-hydroxyneamine alpha-D-glucosyltransferase |
| R11272 | UDP-alpha-D-glucose:neamine alpha-D-glucosyltransferase |
| R11273 | lividamine:oxygen 6'-oxidoreductase |
| R11276 | lividamine:NADP+ 4'-oxidoreductase |
| R11277 | nebramine:NADP+ 4'-oxidoreductase |
| R11278 | UDP-alpha-D-kanosamine:nebramine alpha-D-kanosaminyltransferase |
| R11279 | kanamycin-B:carbamoyl-phosphate ligase (AMP,phosphate-forming) |
| R11280 | paromamine:5-phospho-alpha-D-ribose-1-diphosphate phosphoribosyltransferase |
| R11281 | 5''-phosphoribosylparomamine phosphohydrolase |
| R11282 | UDP-N-acetyl-alpha-D-glucosamine:5-ribosylparomamine N-acetyl-D-glucosaminyltransferase |
| R11283 | 2'''-N-acetyl-6'''-deamino-6'''-hydroxyparomomycin-II hydrolase (acetate-forming) |
| R11284 | 6'''-deamino-6'''-hydroxyparomomycin-II:oxygen 6'''-oxidoreductase |
| R11285 | paromomycin-II:2-oxoglutarate aminotransferase |
| R11286 | paromomycin-II 5'''-epimerase |
| R11288 | 5'''-epi-lividomycin-B 5'''-epimerase |
| R11289 | UDP-D-xylose:paromamine alpha-D-xylosyltransferase |
| R11290 | gentamicin-A2:NAD+ 3''-oxidoreductase |
| R11291 | 3''-dehydro-3''-amino-gentamicin-A2:2-oxoglutarate aminotransferase |
| R11292 | S-adenosyl-L-methionine:3''-dehydro-3''-amino-gentamicin-A2 N-methyltransferase |
| R11293 | S-adenosyl-L-methionine:methylcob(III)alamin:gentamicin-A methyltransferase |
| R11297 | antibiotic-JI-20A:2-oxoglutarate aminotransferase |
| R11298 | antibiotic-JI-20Ba:2-oxoglutarate aminotransferase |
| R11299 | antibiotic-JI-20B:2-oxoglutarate aminotransferase |
| R13180 | S-adenosyl-L-methionine:gentamicin X2 C6'-methyltransferase |
|
|---|
| Compound |
| C00043 | UDP-N-acetyl-alpha-D-glucosamine |
| C17580 | 2-Deoxy-scyllo-inosamine |
| C17581 | 3-Amino-2,3-dideoxy-scyllo-inosose |
| C17583 | 6'-Dehydro-6'-oxoparomamine |
| C17587 | 2'''-N-Acetyl-6'''-deamino-6'''-hydroxyneomycin C |
| C17588 | 6'''-Deamino-6'''-hydroxyneomycin C |
| C17589 | 6'''-Deamino-6'''-oxoneomycin C |
| C18004 | 5''-Phosphoribostamycin |
| C18005 | gamma-L-Glutamyl-butirosin B |
| C18008 | 4-(gamma-L-Glutamylamino)butanoyl-[acp] |
| C18009 | 4-(gamma-L-Glutamylamino)-(2S)-2-hydroxybutanoyl-[acp] |
| C20350 | 2'-Deamino-2'-hydroxyneamine |
| C20351 | 2'-Deamino-2'-hydroxy-6'-dehydroparomamine |
| C20353 | 2'-Deamino-2'-hydroxyparomamine |
| C20699 | 6''-O-Carbamoylkanamycin A |
| C21254 | 3''-Deamino-3''-hydroxykanamycin B |
| C21260 | 5''-Phosphoribosylparomamine |
| C21261 | 2'''-N-Acetyl-6'''-deamino-6'''-hydroxyparomomycin II |
| C21262 | 6'''-Deamino-6'''-hydroxyparomomycin II |
| C21263 | 6'''-Deamino-6'''-oxoparomomycin II |
| C21266 | 3''-Amino-3''-deoxygentamicin A2 |
|
|---|
| Reference |
|
|---|
| Authors |
Kudo F, Eguchi T |
|---|
| Title |
Biosynthetic enzymes for the aminoglycosides butirosin and neomycin. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kudo F, Eguchi T |
|---|
| Title |
Biosynthetic genes for aminoglycoside antibiotics. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Nango E, Kumasaka T, Hirayama T, Tanaka N, Eguchi T |
|---|
| Title |
Structure of 2-deoxy-scyllo-inosose synthase, a key enzyme in the biosynthesis of 2-deoxystreptamine-containing aminoglycoside antibiotics, in complex with a mechanism-based inhibitor and NAD+. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Yokoyama K, Kudo F, Kuwahara M, Inomata K, Tamegai H, Eguchi T, Kakinuma K |
|---|
| Title |
Stereochemical recognition of doubly functional aminotransferase in 2-deoxystreptamine biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Tamegai H, Nango E, Kuwahara M, Yamamoto H, Ota Y, Kuriki H, Eguchi T, Kakinuma K |
|---|
| Title |
Identification of L-glutamine: 2-deoxy-scyllo-inosose aminotransferase required for the biosynthesis of butirosin in Bacillus circulans. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kudo F, Yamamoto Y, Yokoyama K, Eguchi T, Kakinuma K |
|---|
| Title |
Biosynthesis of 2-deoxystreptamine by three crucial enzymes in Streptomyces fradiae NBRC 12773. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Yokoyama K, Ohmori D, Kudo F, Eguchi T |
|---|
| Title |
Mechanistic study on the reaction of a radical SAM dehydrogenase BtrN by electron paramagnetic resonance spectroscopy. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Yokoyama K, Numakura M, Kudo F, Ohmori D, Eguchi T |
|---|
| Title |
Characterization and mechanistic study of a radical SAM dehydrogenase in the biosynthesis of butirosin. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Yokoyama K, Yamamoto Y, Kudo F, Eguchi T |
|---|
| Title |
Involvement of two distinct N-acetylglucosaminyltransferases and a dual-function deacetylase in neomycin biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Fan Q, Huang F, Leadlay PF, Spencer JB |
|---|
| Title |
The neomycin biosynthetic gene cluster of Streptomyces fradiae NCIMB 8233: genetic and biochemical evidence for the roles of two glycosyltransferases and a deacetylase. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Truman AW, Huang F, Llewellyn NM, Spencer JB |
|---|
| Title |
Characterization of the enzyme BtrD from Bacillus circulans and revision of its functional assignment in the biosynthesis of butirosin. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kudo F, Fujii T, Kinoshita S, Eguchi T |
|---|
| Title |
Unique O-ribosylation in the biosynthesis of butirosin. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Yukita T, Nishida H, Eguchi T, Kakinuma K |
|---|
| Title |
Biosynthesis of (2R)-4-amino-2-hydroxybutyric acid, unique and biologically significant substituent in butirosins. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Li Y, Llewellyn NM, Giri R, Huang F, Spencer JB |
|---|
| Title |
Biosynthesis of the unique amino acid side chain of butirosin: possible protective-group chemistry in an acyl carrier protein-mediated pathway. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Llewellyn NM, Li Y, Spencer JB |
|---|
| Title |
Biosynthesis of butirosin: transfer and deprotection of the unique amino acid side chain. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Llewellyn NM, Spencer JB |
|---|
| Title |
Chemoenzymatic acylation of aminoglycoside antibiotics. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Nepal KK, Oh TJ, Sohng JK |
|---|
| Title |
Heterologous production of paromamine in Streptomyces lividans TK24 using kanamycin biosynthetic genes from Streptomyces kanamyceticus ATCC12853. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Park JW, Park SR, Nepal KK, Han AR, Ban YH, Yoo YJ, Kim EJ, Kim EM, Kim D, Sohng JK, Yoon YJ |
|---|
| Title |
Discovery of parallel pathways of kanamycin biosynthesis allows antibiotic manipulation. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Arya DP (ed). |
|---|
| Title |
Aminoglycoside antibiotics: From chemical biology to drug discovery |
|---|
| Journal |
Wiley-Interscience (2007) |
|---|
| Reference |
|
|---|
| Authors |
Park SR, Park JW, Ban YH, Sohng JK, Yoon YJ |
|---|
| Title |
2-Deoxystreptamine-containing aminoglycoside antibiotics: recent advances in the characterization and manipulation of their biosynthetic pathways. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Llewellyn NM, Spencer JB |
|---|
| Title |
Biosynthesis of 2-deoxystreptamine-containing aminoglycoside antibiotics. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kudo F, Eguchi T |
|---|
| Title |
Aminoglycoside Antibiotics: New Insights into the Biosynthetic Machinery of Old Drugs. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Takeishi R, Kudo F, Numakura M, Eguchi T |
|---|
| Title |
Epimerization at C-3'' in butirosin biosynthesis by an NAD(+) -dependent dehydrogenase BtrE and an NADPH-dependent reductase BtrF. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Lv M, Ji X, Zhao J, Li Y, Zhang C, Su L, Ding W, Deng Z, Yu Y, Zhang Q |
|---|
| Title |
Characterization of a C3 Deoxygenation Pathway Reveals a Key Branch Point in Aminoglycoside Biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Clausnitzer D, Piepersberg W, Wehmeier UF |
|---|
| Title |
The oxidoreductases LivQ and NeoQ are responsible for the different 6'-modifications in the aminoglycosides lividomycin and neomycin. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Park JW, Hong JS, Parajuli N, Jung WS, Park SR, Lim SK, Sohng JK, Yoon YJ |
|---|
| Title |
Genetic dissection of the biosynthetic route to gentamicin A2 by heterologous expression of its minimal gene set. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Huang C, Huang F, Moison E, Guo J, Jian X, Duan X, Deng Z, Leadlay PF, Sun Y |
|---|
| Title |
Delineating the biosynthesis of gentamicin x2, the common precursor of the gentamicin C antibiotic complex. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Guo J, Huang F, Huang C, Duan X, Jian X, Leeper F, Deng Z, Leadlay PF, Sun Y |
|---|
| Title |
Specificity and promiscuity at the branch point in gentamicin biosynthesis. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Gu Y, Ni X, Ren J, Gao H, Wang D, Xia H |
|---|
| Title |
Biosynthesis of Epimers C2 and C2a in the Gentamicin C Complex. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kim HJ, McCarty RM, Ogasawara Y, Liu YN, Mansoorabadi SO, LeVieux J, Liu HW |
|---|
| Title |
GenK-catalyzed C-6' methylation in the biosynthesis of gentamicin: isolation and characterization of a cobalamin-dependent radical SAM enzyme. |
|---|
| Journal |
|
|---|
Related
pathway |
| rn00520 | Amino sugar and nucleotide sugar metabolism |
|
|---|
| KO pathway |
|
|---|