| Entry |
|
|---|
| Name |
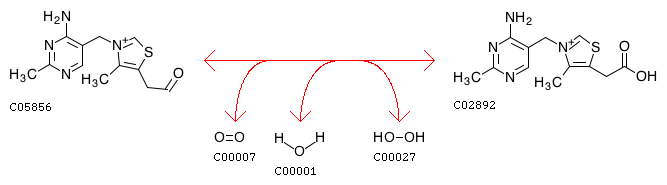
thiamine-aldehyde:oxygen 5-oxidoreductase
|
|---|
| Definition |
Thiamine aldehyde + Oxygen + H2O <=> Hydrogen peroxide + Thiamine acetic acid
|
|---|
| Equation |
|
|---|
|
 |
|---|
| Comment |
|
|---|
| Reaction class |
|
|---|
| Enzyme |
|
|---|
| Pathway |
|
|---|
| Brite |
Enzymatic reactions [BR:br08201]
1. Oxidoreductase reactions
1.1 Acting on the CH-OH group of donors
1.1.3 With oxygen as acceptor
1.1.3.23
R00072 Thiamine aldehyde + Oxygen + H2O <=> Hydrogen peroxide + Thiamine acetic acid
|
|---|
| Reference |
|
|---|
| Authors |
Gomez-Moreno C, Edmondson DE. |
|---|
| Title |
Evidence for an aldehyde intermediate in the catalytic mechanism of thiamine oxidase. |
|---|
| Journal |
|
|---|
| LinkDB |
|
|---|