| Entry |
|
|---|
| Name |
Efferocytosis - Nymphalis io (European peacock) |
|---|
| Class |
Cellular Processes; Transport and catabolism
|
|---|
| Pathway map |
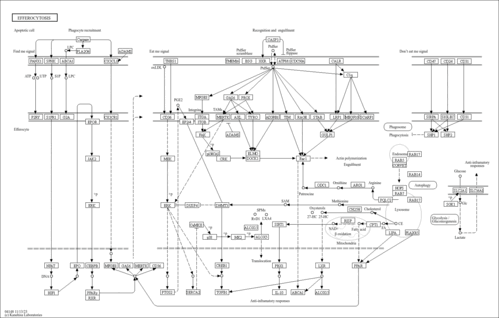
|
|---|
| Organism |
Nymphalis io (European peacock) [GN: niq]
|
|---|
| Gene |
| 126768901 | aryl hydrocarbon receptor nuclear translocator homolog isoform X1 [KO:K09097] |
| 126770133 | cyclic AMP response element-binding protein B isoform X1 [KO:K05870] |
| 126779075 | PTB domain-containing adapter protein ced-6 isoform X1 [KO:K23285] |
| 126774109 | vacuolar protein sorting-associated protein 8 homolog [KO:K20178] |
| 126771578 | vacuolar protein sorting-associated protein 11 homolog [KO:K20179] |
| 126772670 | vacuolar protein sorting-associated protein 16 homolog [KO:K20180] |
| 126770494 | vacuolar protein sorting-associated protein 18 homolog isoform X1 [KO:K20181] |
| 126770464 | vacuolar protein sorting-associated protein 41 homolog [KO:K20184] |
| 126779983 | solute carrier family 2, facilitated glucose transporter member 1-like [KO:K07299] |
|
|---|
| Compound |
| C00019 | S-Adenosyl-L-methionine |
| C04230 | 1-Acyl-sn-glycero-3-phosphocholine |
| C06124 | Sphingosine 1-phosphate |
| C15610 | Cholest-5-ene-3beta,26-diol |
|
|---|
| Reference |
|
|---|
| Authors |
Doran AC, Yurdagul A Jr, Tabas I |
|---|
| Title |
Efferocytosis in health and disease. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Zhang J, Ding W, Zhao M, Liu J, Xu Y, Wan J, Wang M |
|---|
| Title |
Mechanisms of efferocytosis in determining inflammation resolution: Therapeutic potential and the association with cardiovascular disease. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kawano M, Nagata S |
|---|
| Title |
Efferocytosis and autoimmune disease. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Mehrotra P, Ravichandran KS |
|---|
| Title |
Drugging the efferocytosis process: concepts and opportunities. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Park SY, Kim IS |
|---|
| Title |
Engulfment signals and the phagocytic machinery for apoptotic cell clearance. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Kojima Y, Weissman IL, Leeper NJ |
|---|
| Title |
The Role of Efferocytosis in Atherosclerosis. |
|---|
| Journal |
|
|---|
Related
pathway |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|