 | | PATHWAY: rsz00380 | |
| Entry |
|
|---|
| Name |
Tryptophan metabolism - Raphanus sativus (radish) |
|---|
| Class |
Metabolism; Amino acid metabolism
|
|---|
| Pathway map |
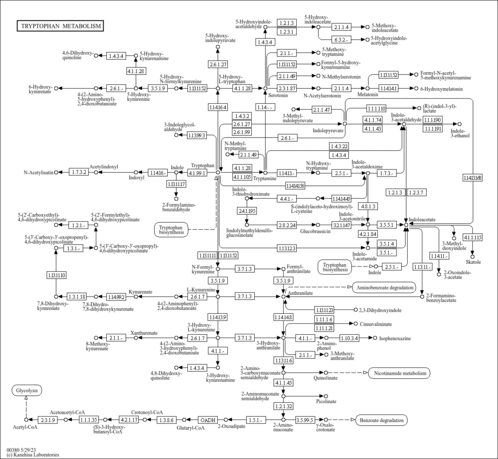
|
|---|
| Module |
|
|---|
| Other DBs |
|
|---|
| Organism |
Raphanus sativus (radish) [GN: rsz]
|
|---|
| Gene |
| 108827241 | dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 1, mitochondrial [KO:K00658] [EC:2.3.1.61] |
| 108848112 | dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 1, mitochondrial [KO:K00658] [EC:2.3.1.61] |
| 108833344 | dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 1, mitochondrial isoform X1 [KO:K00658] [EC:2.3.1.61] |
| 108813150 | LOW QUALITY PROTEIN: aldehyde dehydrogenase family 2 member B4, mitochondrial [KO:K00128] [EC:1.2.1.3] |
| 108842544 | aromatic aminotransferase ISS1 isoform X1 [KO:K00837] [EC:2.6.1.-] |
| 108822742 | 2-oxoglutarate-dependent dioxygenase DAO [KO:K23947] [EC:1.14.11.-] |
| 108857062 | 2-oxoglutarate-dependent dioxygenase DAO-like [KO:K23947] [EC:1.14.11.-] |
| 108837141 | 2-oxoglutarate-dependent dioxygenase DAO-like [KO:K23947] [EC:1.14.11.-] |
|
|---|
| Compound |
| C01111 | 7,8-Dihydroxykynurenate |
| C01144 | (S)-3-Hydroxybutanoyl-CoA |
| C01249 | 7,8-Dihydro-7,8-dihydroxykynurenate |
| C01252 | 4-(2-Aminophenyl)-2,4-dioxobutanoate |
| C01717 | 4-Hydroxy-2-quinolinecarboxylic acid |
| C02161 | 2-Aminophenoxazin-3-one |
| C02937 | Indole-3-acetaldehyde oxime |
| C03230 | (Indol-3-yl)glycolaldehyde |
| C03574 | 2-Formylaminobenzaldehyde |
| C03824 | 2-Aminomuconate semialdehyde |
| C04409 | 2-Amino-3-carboxymuconate semialdehyde |
| C05634 | 5-Hydroxyindoleacetaldehyde |
| C05641 | 5-(3'-Carboxy-3'-oxopropenyl)-4,6-dihydroxypicolinate |
| C05642 | Formyl-N-acetyl-5-methoxykynurenamine |
| C05645 | 4-(2-Amino-3-hydroxyphenyl)-2,4-dioxobutanoate |
| C05646 | 5-Hydroxyindolepyruvate |
| C05647 | Formyl-5-hydroxykynurenamine |
| C05648 | 5-Hydroxy-N-formylkynurenine |
| C05652 | 4-(2-Amino-5-hydroxyphenyl)-2,4-dioxobutanoate |
| C05654 | 5-(2'-Formylethyl)-4,6-dihydroxypicolinate |
| C05655 | 5-(2'-Carboxyethyl)-4,6-dihydroxypicolinate |
| C05656 | 5-(3'-Carboxy-3'-oxopropyl)-4,6-dihydroxypicolinate |
| C05832 | 5-Hydroxyindoleacetylglycine |
| C05835 | 2-Formaminobenzoylacetate |
| C16516 | Indolylmethylthiohydroximate |
| C16517 | Indolylmethyl-desulfoglucosinolate |
| C16518 | S-(Indolylmethylthiohydroximoyl)-L-cysteine |
| C22006 | (R)-(Indol-3-yl)lactate |
|
|---|
| Reference |
|
|---|
| Authors |
Woodward AW, Bartel B |
|---|
| Title |
Auxin: regulation, action, and interaction. |
|---|
| Journal |
|
|---|
Related
pathway |
| rsz00400 | Phenylalanine, tyrosine and tryptophan biosynthesis |
| rsz00760 | Nicotinate and nicotinamide metabolism |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|
|
|