| Entry |
|
|---|
| Definition |
NAD+ + Histone N6-acetyl-L-lysine + H2O <=> Nicotinamide + Histone-L-lysine + O-Acetyl-ADP-ribose + H+
|
|---|
| Equation |
|
|---|
|
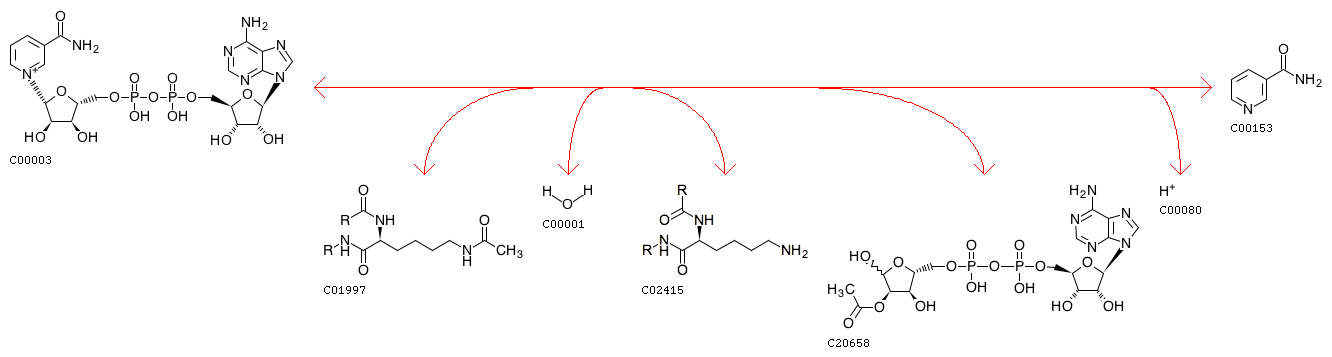 |
|---|
| Reaction class |
|
|---|
| Enzyme |
|
|---|
| Pathway |
| rn00760 | Nicotinate and nicotinamide metabolism |
|
|---|
| Brite |
Enzymatic reactions [BR:br08201]
2. Transferase reactions
2.3 Acyltransferases
2.3.1 Transferring groups other than aminoacyl groups
2.3.1.286
R10633 NAD+ + Histone N6-acetyl-L-lysine + H2O <=> Nicotinamide + Histone-L-lysine + O-Acetyl-ADP-ribose + H+
IUBMB reaction hierarchy [BR:br08202]
2. Transferase reactions
2.3.1.286
R10634 Protein N6-acetyl-L-lysine <=> Protein lysine
R10633 Histone N6-acetyl-L-lysine <=> Histone-L-lysine
|
|---|
| Orthology |
|
|---|
| LinkDB |
|
|---|