 | | PATHWAY: ko00400 | |
| Entry |
|
|---|
| Name |
Phenylalanine, tyrosine and tryptophan biosynthesis |
|---|
| Class |
Metabolism; Amino acid metabolism
|
|---|
| Pathway map |
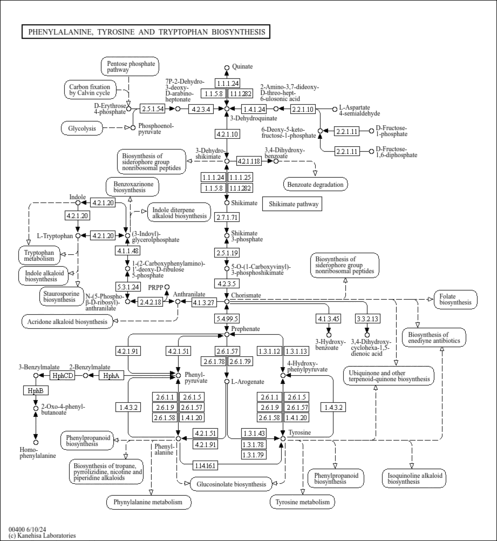
| ko00400 | Phenylalanine, tyrosine and tryptophan biosynthesis |
|
|---|
| Module |
| M00022 | Shikimate pathway, phosphoenolpyruvate + erythrose-4P => chorismate [PATH:ko00400] |
| M00023 | Tryptophan biosynthesis, chorismate => tryptophan [PATH:ko00400] |
| M00024 | Phenylalanine biosynthesis, chorismate => phenylpyruvate => phenylalanine [PATH:ko00400] |
| M00025 | Tyrosine biosynthesis, chorismate => HPP => tyrosine [PATH:ko00400] |
| M00040 | Tyrosine biosynthesis, chorismate => arogenate => tyrosine [PATH:ko00400] |
| M00910 | Phenylalanine biosynthesis, chorismate => arogenate => phenylalanine [PATH:ko00400] |
|
|---|
| Other DBs |
|
|---|
| Orthology |
| K00800 | aroA; 3-phosphoshikimate 1-carboxyvinyltransferase [EC:2.5.1.19] |
| K00811 | ASP5; aspartate aminotransferase, chloroplastic [EC:2.6.1.1] |
| K01626 | E2.5.1.54, aroF, aroG, aroH; 3-deoxy-7-phosphoheptulonate synthase [EC:2.5.1.54] |
| K03856 | AROA2, aroA; 3-deoxy-7-phosphoheptulonate synthase [EC:2.5.1.54] |
| K13498 | trpCF; indole-3-glycerol phosphate synthase / phosphoribosylanthranilate isomerase [EC:4.1.1.48 5.3.1.24] |
| K14454 | GOT1; aspartate aminotransferase, cytoplasmic [EC:2.6.1.1] |
| K14455 | GOT2; aspartate aminotransferase, mitochondrial [EC:2.6.1.1] |
| K17748 | hphA; benzylmalate synthase [EC:2.3.3.-] |
| K17749 | hphCD; 3-benzylmalate isomerase [EC:4.2.1.-] |
| K17750 | hphB; 3-benzylmalate dehydrogenase [EC:1.1.1.-] |
| K24017 | priA; phosphoribosyl isomerase A |
| K24018 | K24018; cyclohexadieny/prephenate dehydrogenase / 3-phosphoshikimate 1-carboxyvinyltransferase |
| K25901 | qsuD; quinate/shikimate dehydrogenase (NAD+) |
| K26400 | quiC1, QA-4; 3-dehydroshikimate dehydratase |
|
|---|
| Compound |
| C00119 | 5-Phospho-alpha-D-ribose 1-diphosphate |
| C00279 | D-Erythrose 4-phosphate |
| C00354 | D-Fructose 1,6-bisphosphate |
| C00441 | L-Aspartate 4-semialdehyde |
| C01179 | 3-(4-Hydroxyphenyl)pyruvate |
| C01269 | 5-O-(1-Carboxyvinyl)-3-phosphoshikimate |
| C01302 | 1-(2-Carboxyphenylamino)-1-deoxy-D-ribulose 5-phosphate |
| C03506 | Indoleglycerol phosphate |
| C04302 | N-(5-Phospho-D-ribosyl)anthranilate |
| C04691 | 2-Dehydro-3-deoxy-D-arabino-heptonate 7-phosphate |
| C16848 | 6-Deoxy-5-ketofructose 1-phosphate |
| C16850 | 2-Amino-3,7-dideoxy-D-threo-hept-6-ulosonic acid |
| C20327 | 2-Oxo-4-phenylbutyric acid |
| C20710 | (4R,5R)-4,5-Dihydroxycyclohexa-1(6),2-diene-1-carboxylate |
|
|---|
Related
pathway |
| ko00010 | Glycolysis / Gluconeogenesis |
| ko00130 | Ubiquinone and other terpenoid-quinone biosynthesis |
| ko00403 | Indole diterpene alkaloid biosynthesis |
| ko00710 | Carbon fixation by Calvin cycle |
| ko00901 | Indole alkaloid biosynthesis |
| ko00940 | Phenylpropanoid biosynthesis |
| ko00950 | Isoquinoline alkaloid biosynthesis |
| ko00960 | Tropane, piperidine and pyridine alkaloid biosynthesis |
| ko01053 | Biosynthesis of siderophore group nonribosomal peptides |
| ko01059 | Biosynthesis of enediyne antibiotics |
|
|---|
|
|