 | | PATHWAY: ko00240 | |
| Entry |
|
|---|
| Name |
Pyrimidine metabolism |
|---|
| Class |
Metabolism; Nucleotide metabolism
|
|---|
| Pathway map |
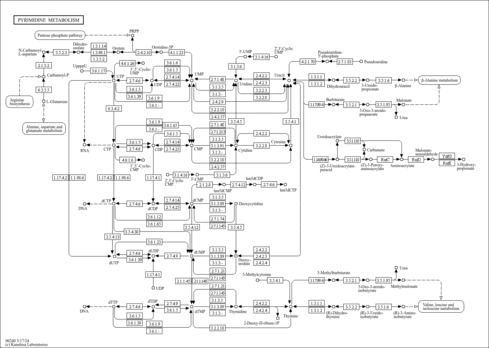
|
|---|
| Module |
| M00046 | Pyrimidine degradation, uracil => beta-alanine, thymine => 3-aminoisobutanoate [PATH:ko00240] |
| M00051 | De novo pyrimidine biosynthesis, glutamine (+ PRPP) => UMP [PATH:ko00240] |
| M00052 | Pyrimidine ribonucleotide biosynthesis, UMP => UDP/UTP,CDP/CTP [PATH:ko00240] |
| M00053 | Deoxyribonucleotide biosynthesis, ADP/GDP/CDP/UDP => dATP/dGTP/dCTP/dUTP [PATH:ko00240] |
| M00938 | Pyrimidine deoxyribonucleotide biosynthesis, UDP => dTTP [PATH:ko00240] |
| M00939 | Pyrimidine degradation, uracil => 3-hydroxypropanoate [PATH:ko00240] |
| M00968 | Pentose bisphosphate pathway (nucleoside degradation), archaea, nucleoside/NMP => 3-PGA/glycerone phosphate [PATH:ko00240] |
|
|---|
| Drug |
| D00323 | Flucytosine (JP18/USP/INN) |
| D00391 | Trifluridine (JAN/USP/INN) |
| D00488 | Pyrimethamine (JAN/USP/INN) |
| D02387 | Cycloguanil pamoate (USAN) |
| D02487 | Proguanil hydrochloride (JAN/USP) |
| D03034 | Azelaic acid (USAN/INN) |
| D12505 | Olorofim (JAN/USAN/INN) |
|
|---|
| Other DBs |
|
|---|
| Orthology |
| K00207 | DPYD; dihydropyrimidine dehydrogenase (NADP+) [EC:1.3.1.2] |
| K00525 | E1.17.4.1A, nrdA, nrdE; ribonucleoside-diphosphate reductase alpha chain [EC:1.17.4.1] |
| K00526 | E1.17.4.1B, nrdB, nrdF; ribonucleoside-diphosphate reductase beta chain [EC:1.17.4.1] |
| K00527 | rtpR; ribonucleoside-triphosphate reductase (thioredoxin) [EC:1.17.4.2] |
| K00609 | pyrB, PYR2; aspartate carbamoyltransferase catalytic subunit [EC:2.1.3.2] |
| K00610 | pyrI; aspartate carbamoyltransferase regulatory subunit |
| K01250 | rihA; pyrimidine-specific ribonucleoside hydrolase [EC:3.2.2.-] |
| K01511 | ENTPD5_6; ectonucleoside triphosphate diphosphohydrolase 5/6 [EC:3.6.1.6] |
| K01513 | ENPP1_3, CD203; ectonucleotide pyrophosphatase/phosphodiesterase family member 1/3 [EC:3.1.4.1 3.6.1.9] |
| K01955 | carB, CPA2; carbamoyl-phosphate synthase large subunit [EC:6.3.5.5] |
| K01956 | carA, CPA1; carbamoyl-phosphate synthase small subunit [EC:6.3.5.5] |
| K02823 | pyrDII; dihydroorotate dehydrogenase electron transfer subunit |
| K02825 | pyrR; pyrimidine operon attenuation protein / uracil phosphoribosyltransferase [EC:2.4.2.9] |
| K06287 | yhdE; nucleoside triphosphate pyrophosphatase [EC:3.6.1.-] |
| K06966 | ppnN; pyrimidine/purine-5'-nucleotide nucleosidase [EC:3.2.2.10 3.2.2.-] |
| K07043 | upp; UTP pyrophosphatase [EC:3.6.1.-] |
| K08723 | yjjG; pyrimidine 5'-nucleotidase [EC:3.1.3.-] |
| K09019 | rutE; 3-hydroxypropanoate dehydrogenase [EC:1.1.1.-] |
| K09021 | rutC; aminoacrylate peracid reductase |
| K09023 | rutD; aminoacrylate hydrolase [EC:3.5.1.-] |
| K09024 | rutF; flavin reductase [EC:1.5.1.-] |
| K09769 | ymdB; 2',3'-cyclic-nucleotide 2'-phosphodiesterase [EC:3.1.4.16] |
| K10807 | RRM1; ribonucleoside-diphosphate reductase subunit M1 [EC:1.17.4.1] |
| K10808 | RRM2; ribonucleoside-diphosphate reductase subunit M2 [EC:1.17.4.1] |
| K12304 | CANT1; soluble calcium-activated nucleotidase 1 [EC:3.6.1.6] |
| K12305 | ENTPD4, LALP70; ectonucleoside triphosphate diphosphohydrolase 4 [EC:3.6.1.6] |
| K12700 | rihC; non-specific riboncleoside hydrolase [EC:3.2.2.-] |
| K16066 | ydfG; 3-hydroxy acid dehydrogenase / malonic semialdehyde reductase [EC:1.1.1.381 1.1.1.-] |
| K17722 | preT; dihydropyrimidine dehydrogenase (NAD+) subunit PreT [EC:1.3.1.1] |
| K17723 | preA; dihydropyrimidine dehydrogenase (NAD+) subunit PreA [EC:1.3.1.1] |
| K17828 | pyrDI; dihydroorotate dehydrogenase (NAD+) catalytic subunit [EC:1.3.1.14] |
| K19797 | gp1; Straboviridae T-even-induced deoxynucleotide kinase [EC:2.7.4.12] |
| K21636 | nrdD; ribonucleoside-triphosphate reductase (formate) [EC:1.1.98.6] |
| K24242 | NT5C3; 7-methylguanosine nucleotidase |
| K24670 | K24670; 2',3'-cyclic-nucleotide 2'-phosphodiesterase / phosphoenolpyruvate phosphatase |
| K25434 | pynA; pyrimidine 5'-nucleotidase |
| K25435 | ASMTL; putative bifunctional nucleoside triphosphate pyrophosphatase / methyltransferase |
| K25589 | inuH; inosine/uridine nucleosidase |
| K26955 | NUDT25, NUDX25; bis(5'-nucleosidyl)-tetraphosphatase |
| K27603 | pycC; uridylate cyclase |
| K27604 | pycC; cytidylate cyclase |
|
|---|
| Compound |
| C00119 | 5-Phospho-alpha-D-ribose 1-diphosphate |
| C00438 | N-Carbamoyl-L-aspartate |
| C00672 | 2-Deoxy-D-ribose 1-phosphate |
| C01168 | Pseudouridine 5'-phosphate |
| C01205 | (R)-3-Amino-2-methylpropanoate |
| C03997 | 5-Hydroxymethyldeoxycytidylate |
| C06198 | P1,P4-Bis(5'-uridyl) tetraphosphate |
| C11038 | 2'-Deoxy-5-hydroxymethylcytidine-5'-diphosphate |
| C11039 | 2'-Deoxy-5-hydroxymethylcytidine-5'-triphosphate |
| C15607 | 3-Oxo-3-ureidopropanoate |
| C20231 | (Z)-3-Ureidoacrylate peracid |
| C20249 | (Z)-3-Peroxyaminoacrylate |
| C20846 | 3-Oxo-3-ureidoisobutyrate |
| C21029 | (R)-3-Ureidoisobutyrate |
|
|---|
Related
pathway |
| ko00250 | Alanine, aspartate and glutamate metabolism |
| ko00280 | Valine, leucine and isoleucine degradation |
|
|---|
|
|