| Entry |
|
|---|
| Name |
Basal transcription factors - Malus sylvestris (European crab apple) |
|---|
| Class |
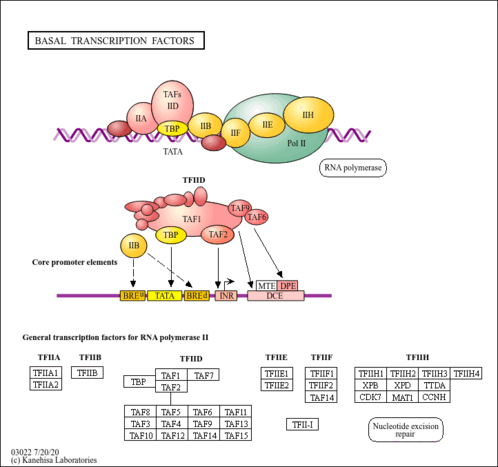
Genetic Information Processing; Transcription
|
|---|
| Pathway map |
|
|---|
| Other DBs |
|
|---|
| Organism |
Malus sylvestris (European crab apple) [GN: msyl]
|
|---|
| Gene |
| 126616825 | transcription initiation factor IIA large subunit-like isoform X1 [KO:K03122] |
| 126589878 | transcription initiation factor IIA large subunit-like [KO:K03122] |
| 126626007 | transcription initiation factor TFIID subunit 2 isoform X1 [KO:K03128] |
| 126630919 | transcription initiation factor TFIID subunit 10-like [KO:K03134] |
| 126603703 | transcription initiation factor TFIID subunit 10-like [KO:K03134] |
| 126632892 | transcription initiation factor TFIID subunit 5 isoform X1 [KO:K03130] |
| 126626401 | transcription initiation factor TFIID subunit 4b isoform X1 [KO:K03129] |
| 126614860 | transcription initiation factor TFIID subunit 4b-like isoform X1 [KO:K03129] |
| 126600787 | transcription initiation factor TFIID subunit 12-like isoform X1 [KO:K03126] |
| 126593771 | transcription initiation factor TFIID subunit 12-like [KO:K03126] |
| 126610389 | transcription initiation factor TFIID subunit 12b-like [KO:K03126] |
| 126583797 | transcription initiation factor TFIID subunit 12b-like [KO:K03126] |
| 126609914 | transcription initiation factor TFIID subunit 6-like isoform X1 [KO:K03131] |
| 126626693 | transcription initiation factor IIE subunit beta-like [KO:K03137] |
| 126586200 | transcription initiation factor IIF subunit alpha-like [KO:K03138] |
| 126623951 | transcription initiation factor IIF subunit alpha-like [KO:K03138] |
| 126633160 | transcription initiation factor IIF subunit beta-like [KO:K03139] [EC:5.6.2.-] |
| 126605353 | transcription initiation factor IIF subunit beta-like [KO:K03139] [EC:5.6.2.-] |
| 126601677 | general transcription and DNA repair factor IIH subunit TFB1-3-like isoform X1 [KO:K03141] |
| 126632855 | general transcription and DNA repair factor IIH subunit TFB1-3-like isoform X1 [KO:K03141] |
| 126590830 | general transcription factor IIH subunit 2-like isoform X1 [KO:K03142] |
| 126603951 | general transcription and DNA repair factor IIH subunit TFB4 [KO:K03143] |
| 126587213 | general transcription and DNA repair factor IIH subunit TFB2-like isoform X1 [KO:K03144] |
| 126622368 | general transcription and DNA repair factor IIH subunit TFB2-like [KO:K03144] |
| 126626938 | general transcription and DNA repair factor IIH helicase subunit XPB1 isoform X1 [KO:K10843] [EC:5.6.2.4] |
| 126620178 | general transcription and DNA repair factor IIH subunit TFB5 [KO:K10845] |
| 126594349 | general transcription and DNA repair factor IIH subunit TFB5 [KO:K10845] |
|
|---|
| Reference |
|
|---|
| Authors |
Thomas MC, Chiang CM |
|---|
| Title |
The general transcription machinery and general cofactors. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Martinez E |
|---|
| Title |
Multi-protein complexes in eukaryotic gene transcription. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Cler E, Papai G, Schultz P, Davidson I |
|---|
| Title |
Recent advances in understanding the structure and function of general transcription factor TFIID. |
|---|
| Journal |
|
|---|
| Reference |
|
|---|
| Authors |
Malik S, Roeder RG. |
|---|
| Title |
Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. |
|---|
| Journal |
|
|---|
Related
pathway |
|
|---|
| KO pathway |
|
|---|
| LinkDB |
|
|---|