| Entry |
|
|---|
| Definition |
|
|---|
|
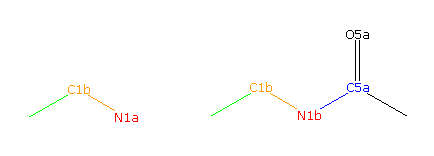 |
|---|
| Reactant pair |
|
|---|
| Related class |
|
|---|
| Reaction |
|
|---|
| Enzyme |
|
|---|
| Pathway |
| rn00120 | Primary bile acid biosynthesis |
| rn00260 | Glycine, serine and threonine metabolism |
| rn00330 | Arginine and proline metabolism |
| rn00430 | Taurine and hypotaurine metabolism |
| rn00440 | Phosphonate and phosphinate metabolism |
| rn00524 | Neomycin, kanamycin and gentamicin biosynthesis |
| rn00770 | Pantothenate and CoA biosynthesis |
| rn00975 | Biosynthesis of various siderophores |
| rn00983 | Drug metabolism - other enzymes |
| rn00996 | Biosynthesis of various alkaloids |
| rn00998 | Biosynthesis of various antibiotics |
| rn00999 | Biosynthesis of various plant secondary metabolites |
| rn01110 | Biosynthesis of secondary metabolites |
| rn01120 | Microbial metabolism in diverse environments |
| rn01230 | Biosynthesis of amino acids |
|
|---|
| RModule |
| RM024 | Pyrimidine degradation |
|
|---|
| Orthology |
| K01270 | dipeptidase D [EC:3.4.13.-] |
| K02225 | cobalamin biosynthesis protein CobC |
| K03524 | BirA family transcriptional regulator, biotin operon repressor / biotin---[acetyl-CoA-carboxylase] ligase [EC:6.3.4.15] |
| K11693 | peptidoglycan pentaglycine glycine transferase (the first glycine) [EC:2.3.2.16] |
| K11694 | peptidoglycan pentaglycine glycine transferase (the second and third glycine) [EC:2.3.2.17] |
| K11695 | peptidoglycan pentaglycine glycine transferase (the fourth and fifth glycine) [EC:2.3.2.18] |
| K13564 | gamma-L-glutamyl-butirosin B gamma-L-glutamyl cyclotransferase [EC:4.3.2.6] |
| K15428 | Cys-Gly metallodipeptidase DUG1 [EC:3.4.13.-] |
| K18566 | N-acetyltransferase [EC:2.3.1.-] |
| K18572 | putative pantetheine hydrolase [EC:3.5.1.-] |
|
|---|